You are browsing environment: FUNGIDB
CAZyme Information: EPrPVT00000021447-p1
You are here: Home > Sequence: EPrPVT00000021447-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytopythium vexans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Pythiaceae; Phytopythium; Phytopythium vexans | |||||||||||
| CAZyme ID | EPrPVT00000021447-p1 | |||||||||||
| CAZy Family | GH78 | |||||||||||
| CAZyme Description | General transcription factor IIH subunit | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH17 | 41 | 294 | 2.9e-22 | 0.9453376205787781 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 237285 | trpA | 7.47e-108 | 723 | 950 | 1 | 228 | tryptophan synthase subunit alpha; Provisional |
| 240075 | Tryptophan_synthase_alpha | 2.93e-98 | 735 | 950 | 1 | 215 | Ttryptophan synthase (TRPS) alpha subunit (TSA). TPRS is a bifunctional tetrameric enzyme (2 alpha and 2 beta subunits) that catalyzes the last two steps of L-tryptophan biosynthesis. Alpha and beta subunit catalyze two distinct reactions which are both strongly stimulated by the formation of the complex. The alpha subunit catalyzes the cleavage of indole 3-glycerol phosphate (IGP) to indole and d-glyceraldehyde 3-phosphate (G3P). Indole is then channeled to the active site of the beta subunit, a PLP-dependent enzyme that catalyzes a replacement reaction to convert L-serine into L-tryptophan. |
| 223237 | TrpA | 1.59e-93 | 721 | 950 | 4 | 233 | Tryptophan synthase alpha chain [Amino acid transport and metabolism]. |
| 397778 | Tfb4 | 7.27e-92 | 415 | 671 | 3 | 273 | Transcription factor Tfb4. This family appears to be distantly related to the VWA domain. |
| 395227 | Trp_syntA | 2.48e-89 | 725 | 950 | 1 | 226 | Tryptophan synthase alpha chain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 4.39e-74 | 11 | 306 | 10 | 304 | |
| 9.73e-72 | 5 | 304 | 5 | 310 | |
| 3.50e-71 | 16 | 304 | 19 | 313 | |
| 1.11e-70 | 2 | 306 | 1 | 315 | |
| 3.57e-67 | 8 | 303 | 1 | 313 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.85e-67 | 721 | 950 | 4 | 233 | Lbcats [synthetic construct],5EY5_C Lbcats [synthetic construct] |
|
| 3.13e-55 | 415 | 690 | 10 | 305 | Human core TFIIH bound to DNA within the PIC [Homo sapiens],5IY6_3 Human holo-PIC in the closed state [Homo sapiens],5IY7_3 Chain 3, General transcription factor IIH subunit 3 [Homo sapiens],5IY8_3 Human holo-PIC in the initial transcribing state [Homo sapiens],5IY9_3 Human holo-PIC in the initial transcribing state (no IIS) [Homo sapiens],5O85_A p34-p44 complex [Homo sapiens],5O85_C p34-p44 complex [Homo sapiens],5OF4_F Chain F, General transcription factor IIH subunit 3 [Homo sapiens],6NMI_F Cryo-EM structure of the human TFIIH core complex [Homo sapiens],6O9L_4 Human holo-PIC in the closed state [Homo sapiens],6O9M_4 Structure of the human apo TFIIH [Homo sapiens],6RO4_E Structure of the core TFIIH-XPA-DNA complex [Homo sapiens],7AD8_E Chain E, General transcription factor IIH subunit 3 [Homo sapiens],7EGB_3 Chain 3, General transcription factor IIH subunit 3 [Homo sapiens],7EGC_3 Chain 3, General transcription factor IIH subunit 3 [Homo sapiens],7ENA_3 Chain 3, General transcription factor IIH subunit 3 [Homo sapiens],7ENC_3 Chain 3, General transcription factor IIH subunit 3 [Homo sapiens],7LBM_b Chain b, General transcription factor IIH subunit 3 [Homo sapiens],7NVR_4 Chain 4, General transcription factor IIH subunit 3 [Homo sapiens],7NVW_4 Chain 4, General transcription factor IIH subunit 3 [Homo sapiens],7NVX_4 Chain 4, General transcription factor IIH subunit 3 [Homo sapiens],7NVY_4 Chain 4, General transcription factor IIH subunit 3 [Homo sapiens],7NVZ_4 Chain 4, General transcription factor IIH subunit 3 [Homo sapiens],7NW0_4 Chain 4, General transcription factor IIH subunit 3 [Homo sapiens] |
|
| 4.09e-49 | 721 | 950 | 7 | 236 | Crystal structure of Tryptophan synthase alpha chain from Legionella pneumophila subsp. pneumophila [Legionella pneumophila subsp. pneumophila str. Philadelphia 1] |
|
| 1.04e-48 | 721 | 950 | 7 | 236 | Crystal structure of tryptophan synthase subunit alpha from Legionella pneumophila str. Paris [Legionella pneumophila str. Paris] |
|
| 1.29e-46 | 735 | 950 | 6 | 220 | Chain A, Tryptophan synthase alpha chain [Pyrococcus furiosus],2DZP_B Chain B, Tryptophan synthase alpha chain [Pyrococcus furiosus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.67e-65 | 721 | 950 | 4 | 234 | Tryptophan synthase OS=Neurospora crassa (strain ATCC 24698 / 74-OR23-1A / CBS 708.71 / DSM 1257 / FGSC 987) OX=367110 GN=trp-3 PE=1 SV=1 |
|
| 9.15e-65 | 719 | 950 | 3 | 235 | Tryptophan synthase OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=TRP5 PE=1 SV=1 |
|
| 5.00e-62 | 721 | 948 | 4 | 231 | Tryptophan synthase alpha chain OS=Herpetosiphon aurantiacus (strain ATCC 23779 / DSM 785 / 114-95) OX=316274 GN=trpA PE=3 SV=1 |
|
| 6.54e-61 | 721 | 949 | 4 | 232 | Tryptophan synthase alpha chain OS=Roseiflexus castenholzii (strain DSM 13941 / HLO8) OX=383372 GN=trpA PE=3 SV=1 |
|
| 1.24e-60 | 721 | 949 | 4 | 232 | Tryptophan synthase alpha chain OS=Roseiflexus sp. (strain RS-1) OX=357808 GN=trpA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
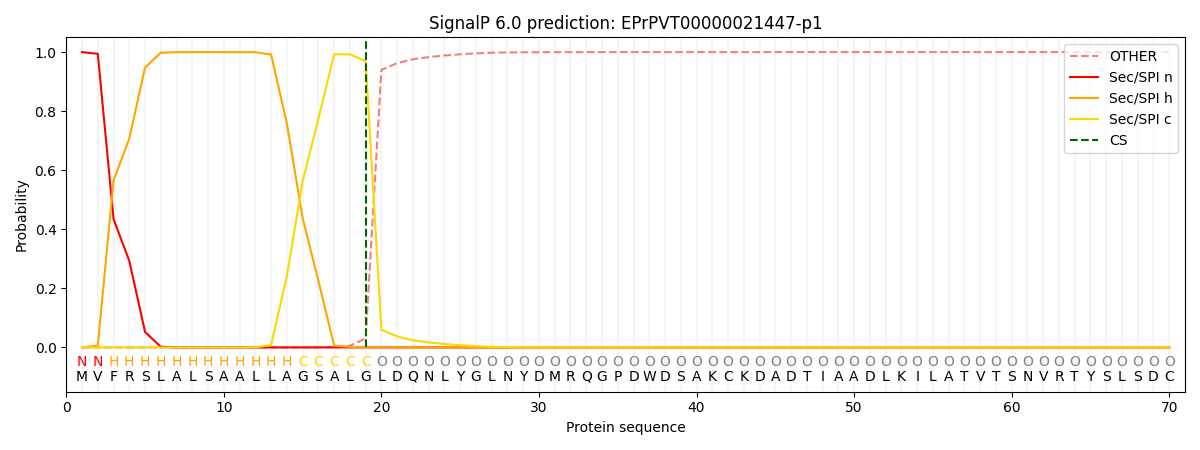
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000380 | 0.999592 | CS pos: 19-20. Pr: 0.9692 |
