You are browsing environment: FUNGIDB
CAZyme Information: EPrPVT00000018144-p1
You are here: Home > Sequence: EPrPVT00000018144-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytopythium vexans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Pythiaceae; Phytopythium; Phytopythium vexans | |||||||||||
| CAZyme ID | EPrPVT00000018144-p1 | |||||||||||
| CAZy Family | GH28 | |||||||||||
| CAZyme Description | Maltose O-acetyltransferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA1 | 86 | 616 | 1.3e-30 | 0.9664804469273743 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 100047 | LbH_MAT_GAT | 1.39e-70 | 657 | 793 | 1 | 169 | Maltose O-acetyltransferase (MAT) and Galactoside O-acetyltransferase (GAT): MAT and GAT catalyze the CoA-dependent acetylation of the 6-hydroxyl group of their respective sugar substrates. MAT acetylates maltose and glucose exclusively at the C6 position of the nonreducing end glucosyl moiety. GAT specifically acetylates galactopyranosides. Furthermore, MAT shows higher affinity toward artificial substrates containing an alkyl or hydrophobic chain as well as a glucosyl unit. Active MAT and GAT are homotrimers, with each subunit consisting of an N-terminal alpha-helical region and a C-terminal left-handed parallel alpha-helix (LbH) subdomain with 6 turns, each containing three imperfect tandem repeats of a hexapeptide repeat motif (X-[STAV]-X-[LIV]-[GAED]-X). |
| 259922 | CuRO_1_Tth-MCO_like | 1.45e-59 | 65 | 189 | 1 | 138 | The first cupredoxin domain of the bacterial laccases similar to Tth-MCO from Thermus Thermophilus. The subfamily of bacterial laccases includes Tth-MCO and similar proteins. Tth-MCO is a hyperthermophilic multicopper oxidase (MCO) from thermus thermophilus HB27. Laccase is a blue multi-copper enzyme that catalyzes the oxidation of a variety aromatic - notably phenolic and inorganic substances coupled to the reduction of molecular oxygen to water. It has been implicated in a wide spectrum of biological activities and, in particular, plays a key role in morphogenesis, development and lignin metabolism in fungi and plants. Although MCOs have diverse functions, majority of them have three cupredoxin domain repeats that include one mononuclear and one trinuclear copper center. The copper ions are bound in several sites: Type 1, Type 2, and/or Type 3. The ensemble of types 2 and 3 copper is called a trinuclear cluster. MCOs oxidize their substrate by accepting electrons at a mononuclear copper center and transferring them to the active site trinuclear copper center. The cupredoxin domain 1 of 3-domain MCOs contains part the trinuclear copper binding site, which is located at the interface of domains 1 and 3. |
| 182235 | PRK10092 | 6.07e-53 | 645 | 796 | 1 | 183 | maltose O-acetyltransferase; Provisional |
| 100053 | LbH_MAT_like | 4.84e-42 | 700 | 793 | 9 | 109 | Maltose O-acyltransferase (MAT)-like: This family is composed of maltose O-acetyltransferase, galactoside O-acetyltransferase (GAT), xenobiotic acyltransferase (XAT) and similar proteins. MAT and GAT catalyze the CoA-dependent acetylation of the 6-hydroxyl group of their respective sugar substrates. MAT acetylates maltose and glucose exclusively while GAT specifically acetylates galactopyranosides. XAT catalyzes the CoA-dependent acetylation of a variety of hydroxyl-bearing acceptors such as chloramphenicol and streptogramin, among others. XATs are implicated in inactivating xenobiotics leading to xenobiotic resistance in patients. Members of this family contain a a left-handed parallel beta-helix (LbH) domain with at least 5 turns, each containing three imperfect tandem repeats of a hexapeptide repeat motif (X-[STAV]-X-[LIV]-[GAED]-X). They are trimeric in their active form. |
| 225043 | SufI | 9.86e-38 | 47 | 617 | 14 | 443 | Multicopper oxidase with three cupredoxin domains (includes cell division protein FtsP and spore coat protein CotA) [Cell cycle control, cell division, chromosome partitioning, Inorganic ion transport and metabolism, Cell wall/membrane/envelope biogenesis]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.85e-41 | 120 | 623 | 5 | 469 | |
| 8.89e-18 | 96 | 615 | 20 | 468 | |
| 6.60e-12 | 90 | 205 | 107 | 217 | |
| 3.08e-10 | 85 | 377 | 45 | 275 | |
| 4.07e-10 | 90 | 210 | 94 | 212 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.57e-34 | 645 | 796 | 1 | 183 | Crystal structure of Maltose O-acetyltransferase from E. coli [Escherichia coli K-12],6AG8_C Crystal structure of Maltose O-acetyltransferase from E. coli [Escherichia coli K-12] |
|
| 5.70e-34 | 647 | 796 | 2 | 182 | E. coli maltose-O-acetyltransferase [Escherichia coli],1OCX_B E. coli maltose-O-acetyltransferase [Escherichia coli],1OCX_C E. coli maltose-O-acetyltransferase [Escherichia coli] |
|
| 1.38e-32 | 645 | 794 | 1 | 182 | Crystal Structure of Maltose Transacetylase from Geobacillus kaustophilus [Geobacillus kaustophilus],2IC7_B Crystal Structure of Maltose Transacetylase from Geobacillus kaustophilus [Geobacillus kaustophilus],2IC7_C Crystal Structure of Maltose Transacetylase from Geobacillus kaustophilus [Geobacillus kaustophilus],2P2O_A Crystal structure of maltose transacetylase from Geobacillus kaustophilus P2(1) crystal form [Geobacillus kaustophilus HTA426],2P2O_B Crystal structure of maltose transacetylase from Geobacillus kaustophilus P2(1) crystal form [Geobacillus kaustophilus HTA426],2P2O_C Crystal structure of maltose transacetylase from Geobacillus kaustophilus P2(1) crystal form [Geobacillus kaustophilus HTA426],2P2O_D Crystal structure of maltose transacetylase from Geobacillus kaustophilus P2(1) crystal form [Geobacillus kaustophilus HTA426],2P2O_E Crystal structure of maltose transacetylase from Geobacillus kaustophilus P2(1) crystal form [Geobacillus kaustophilus HTA426],2P2O_F Crystal structure of maltose transacetylase from Geobacillus kaustophilus P2(1) crystal form [Geobacillus kaustophilus HTA426] |
|
| 4.15e-30 | 642 | 797 | 1 | 189 | Crystal Structure of Maltose O-acetyltransferase from Bacillus anthracis [Bacillus anthracis],3HJJ_B Crystal Structure of Maltose O-acetyltransferase from Bacillus anthracis [Bacillus anthracis],3HJJ_C Crystal Structure of Maltose O-acetyltransferase from Bacillus anthracis [Bacillus anthracis],3IGJ_A Crystal Structure of Maltose O-acetyltransferase Complexed with Acetyl Coenzyme A from Bacillus anthracis [Bacillus anthracis],3IGJ_B Crystal Structure of Maltose O-acetyltransferase Complexed with Acetyl Coenzyme A from Bacillus anthracis [Bacillus anthracis],3IGJ_C Crystal Structure of Maltose O-acetyltransferase Complexed with Acetyl Coenzyme A from Bacillus anthracis [Bacillus anthracis] |
|
| 4.64e-29 | 647 | 797 | 5 | 187 | Chain A, Maltose O-acetyltransferase [Clostridioides difficile 630],3SRT_B Chain B, Maltose O-acetyltransferase [Clostridioides difficile 630],4ISX_A Chain A, Maltose O-acetyltransferase [Clostridioides difficile 630],4ISX_B Chain B, Maltose O-acetyltransferase [Clostridioides difficile 630] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.02e-34 | 645 | 796 | 2 | 184 | Probable maltose O-acetyltransferase OS=Bacillus subtilis (strain 168) OX=224308 GN=maa PE=3 SV=1 |
|
| 6.40e-34 | 645 | 796 | 1 | 183 | Maltose O-acetyltransferase OS=Escherichia coli (strain K12) OX=83333 GN=maa PE=1 SV=3 |
|
| 1.14e-31 | 647 | 796 | 7 | 186 | Putative acetyltransferase DDB_G0280825 OS=Dictyostelium discoideum OX=44689 GN=DDB_G0280825 PE=3 SV=1 |
|
| 9.99e-29 | 643 | 796 | 2 | 185 | Putative acetyltransferase DDB_G0275913 OS=Dictyostelium discoideum OX=44689 GN=DDB_G0275913 PE=3 SV=1 |
|
| 4.13e-26 | 655 | 802 | 10 | 189 | Putative acetyltransferase YJL218W OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=YJL218W PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
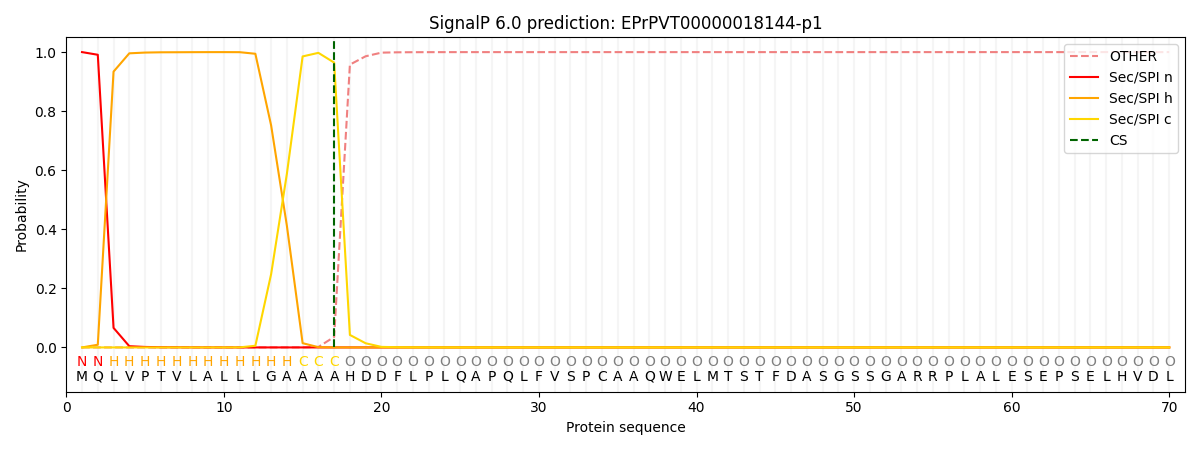
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000282 | 0.999691 | CS pos: 17-18. Pr: 0.9645 |
