You are browsing environment: FUNGIDB
CAZyme Information: EPrPVT00000017929-p1
You are here: Home > Sequence: EPrPVT00000017929-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytopythium vexans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Pythiaceae; Phytopythium; Phytopythium vexans | |||||||||||
| CAZyme ID | EPrPVT00000017929-p1 | |||||||||||
| CAZy Family | GH19 | |||||||||||
| CAZyme Description | Beta-glucosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.21:85 | 3.2.1.62:12 | 3.2.1.118:11 | 3.2.1.149:10 | 3.2.1.105:6 | 3.2.1.117:6 | 3.2.1.161:2 | 2.4.1.-:2 | 3.2.1.38:2 | 3.2.1.206:2 | 3.2.1.186:2 | 3.2.1.-:1 | 3.2.1.125:1 | 3.2.1.119:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH1 | 31 | 418 | 1.1e-109 | 0.7832167832167832 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395176 | Glyco_hydro_1 | 5.05e-118 | 32 | 418 | 5 | 362 | Glycosyl hydrolase family 1. |
| 225343 | BglB | 2.48e-99 | 32 | 335 | 4 | 300 | Beta-glucosidase/6-phospho-beta-glucosidase/beta-galactosidase [Carbohydrate transport and metabolism]. |
| 215435 | PLN02814 | 9.46e-84 | 32 | 364 | 28 | 354 | beta-glucosidase |
| 215455 | PLN02849 | 2.74e-82 | 32 | 372 | 30 | 363 | beta-glucosidase |
| 215539 | PLN02998 | 1.01e-75 | 32 | 344 | 31 | 344 | beta-glucosidase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 4.36e-153 | 2 | 418 | 4 | 416 | |
| 4.53e-152 | 31 | 418 | 22 | 406 | |
| 1.32e-151 | 24 | 418 | 5 | 397 | |
| 6.08e-108 | 80 | 418 | 5 | 329 | |
| 5.28e-86 | 32 | 418 | 39 | 419 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.88e-81 | 32 | 361 | 43 | 366 | Structure of inactive mutant of Strictosidine Glucosidase in complex with strictosidine [Rauvolfia serpentina],2JF6_B Structure of inactive mutant of Strictosidine Glucosidase in complex with strictosidine [Rauvolfia serpentina],2JF7_A Structure of Strictosidine Glucosidase [Rauvolfia serpentina],2JF7_B Structure of Strictosidine Glucosidase [Rauvolfia serpentina],3ZJ7_A Crystal structure of strictosidine glucosidase in complex with inhibitor-1 [Rauvolfia serpentina],3ZJ7_B Crystal structure of strictosidine glucosidase in complex with inhibitor-1 [Rauvolfia serpentina],3ZJ8_A Crystal structure of strictosidine glucosidase in complex with inhibitor-2 [Rauvolfia serpentina],3ZJ8_B Crystal structure of strictosidine glucosidase in complex with inhibitor-2 [Rauvolfia serpentina] |
|
| 1.64e-79 | 32 | 334 | 31 | 323 | Chain A, Glycoside hydrolase [Nannochloris],5YJ7_B Chain B, Glycoside hydrolase [Nannochloris],5YJ7_C Chain C, Glycoside hydrolase [Nannochloris],5YJ7_D Chain D, Glycoside hydrolase [Nannochloris] |
|
| 6.89e-79 | 32 | 336 | 20 | 322 | Crystal Structure of Rice BGlu1 E386S Mutant [Oryza sativa Japonica Group],3SCR_B Crystal Structure of Rice BGlu1 E386S Mutant [Oryza sativa Japonica Group],3SCS_A Crystal Structure of Rice BGlu1 E386S Mutant Complexed with alpha-Glucosyl Fluoride [Oryza sativa Japonica Group],3SCS_B Crystal Structure of Rice BGlu1 E386S Mutant Complexed with alpha-Glucosyl Fluoride [Oryza sativa Japonica Group] |
|
| 6.89e-79 | 32 | 336 | 20 | 322 | Rice BGlu1 beta-glucosidase, a plant exoglucanase/beta-glucosidase [Oryza sativa Japonica Group],2RGL_B Rice BGlu1 beta-glucosidase, a plant exoglucanase/beta-glucosidase [Oryza sativa Japonica Group],2RGM_A Rice BGlu1 beta-glucosidase, a plant exoglucanase/beta-glucosidase [Oryza sativa Japonica Group],2RGM_B Rice BGlu1 beta-glucosidase, a plant exoglucanase/beta-glucosidase [Oryza sativa Japonica Group],7BZM_A Crystal structure of rice Os3BGlu7 with glucoimidazole [Oryza sativa Japonica Group],7BZM_B Crystal structure of rice Os3BGlu7 with glucoimidazole [Oryza sativa Japonica Group] |
|
| 6.89e-79 | 32 | 336 | 20 | 322 | Crystal Structure of Rice BGlu1 E386G/S334A Mutant Complexed with Cellotetraose [Oryza sativa Japonica Group],3SCV_B Crystal Structure of Rice BGlu1 E386G/S334A Mutant Complexed with Cellotetraose [Oryza sativa Japonica Group] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.95e-82 | 32 | 336 | 74 | 378 | Furcatin hydrolase OS=Viburnum furcatum OX=237940 PE=1 SV=1 |
|
| 4.47e-82 | 32 | 418 | 40 | 419 | Beta-glucosidase 17 OS=Arabidopsis thaliana OX=3702 GN=BGLU17 PE=2 SV=1 |
|
| 2.00e-80 | 32 | 361 | 43 | 366 | Strictosidine-O-beta-D-glucosidase OS=Rauvolfia serpentina OX=4060 GN=SGR1 PE=1 SV=1 |
|
| 3.88e-79 | 32 | 336 | 25 | 331 | Beta-glucosidase 25 OS=Oryza sativa subsp. japonica OX=39947 GN=BGLU25 PE=2 SV=2 |
|
| 1.64e-78 | 32 | 418 | 33 | 413 | Beta-glucosidase 24 OS=Oryza sativa subsp. japonica OX=39947 GN=BGLU24 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
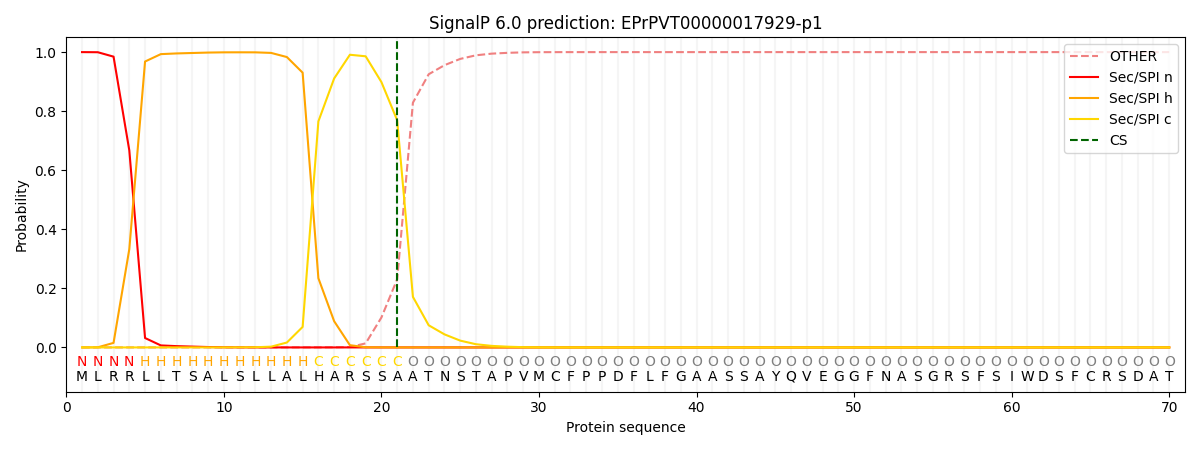
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.001126 | 0.998858 | CS pos: 21-22. Pr: 0.7693 |
