You are browsing environment: FUNGIDB
CAZyme Information: EPrPVT00000016885-p1
You are here: Home > Sequence: EPrPVT00000016885-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytopythium vexans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Pythiaceae; Phytopythium; Phytopythium vexans | |||||||||||
| CAZyme ID | EPrPVT00000016885-p1 | |||||||||||
| CAZy Family | GH16 | |||||||||||
| CAZyme Description | Cellulose synthase 1 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.12:3 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT2 | 585 | 790 | 5.7e-17 | 0.7715736040609137 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 133043 | CESA_CelA_like | 1.73e-44 | 321 | 712 | 1 | 234 | CESA_CelA_like are involved in the elongation of the glucan chain of cellulose. Family of proteins related to Agrobacterium tumefaciens CelA and Gluconacetobacter xylinus BscA. These proteins are involved in the elongation of the glucan chain of cellulose, an aggregate of unbranched polymers of beta-1,4-linked glucose residues. They are putative catalytic subunit of cellulose synthase, which is a glycosyltransferase using UDP-glucose as the substrate. The catalytic subunit is an integral membrane protein with 6 transmembrane segments and it is postulated that the protein is anchored in the membrane at the N-terminal end. |
| 236918 | bcsA | 1.08e-18 | 319 | 798 | 258 | 569 | cellulose synthase catalytic subunit; Provisional |
| 224136 | BcsA | 1.73e-14 | 273 | 782 | 10 | 346 | Glycosyltransferase, catalytic subunit of cellulose synthase and poly-beta-1,6-N-acetylglucosamine synthase [Cell motility]. |
| 404513 | Glyco_trans_2_3 | 1.94e-12 | 545 | 777 | 12 | 193 | Glycosyl transferase family group 2. Members of this family of prokaryotic proteins include putative glucosyltransferases, which are involved in bacterial capsule biosynthesis. |
| 275388 | PH | 5.74e-10 | 75 | 176 | 2 | 90 | Pleckstrin homology (PH) domain. PH domains have diverse functions, but in general are involved in targeting proteins to the appropriate cellular location or in the interaction with a binding partner. They share little sequence conservation, but all have a common fold, which is electrostatically polarized. Less than 10% of PH domains bind phosphoinositide phosphates (PIPs) with high affinity and specificity. PH domains are distinguished from other PIP-binding domains by their specific high-affinity binding to PIPs with two vicinal phosphate groups: PtdIns(3,4)P2, PtdIns(4,5)P2 or PtdIns(3,4,5)P3 which results in targeting some PH domain proteins to the plasma membrane. A few display strong specificity in lipid binding. Any specificity is usually determined by loop regions or insertions in the N-terminus of the domain, which are not conserved across all PH domains. PH domains are found in cellular signaling proteins such as serine/threonine kinase, tyrosine kinases, regulators of G-proteins, endocytotic GTPases, adaptors, as well as cytoskeletal associated molecules and in lipid associated enzymes. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 962 | 1 | 1026 | |
| 0.0 | 1 | 962 | 1 | 1026 | |
| 0.0 | 1 | 962 | 1 | 1028 | |
| 0.0 | 10 | 962 | 13 | 1026 | |
| 0.0 | 11 | 962 | 5 | 1019 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.11e-19 | 313 | 882 | 121 | 532 | Chain A, Putative cellulose synthase [Cereibacter sphaeroides 2.4.1] |
|
| 5.67e-19 | 313 | 882 | 133 | 544 | Chain A, Cellulose Synthase Subunit A [Cereibacter sphaeroides] |
|
| 5.68e-19 | 313 | 882 | 134 | 545 | Chain A, Cellulose Synthase A subunit [Cereibacter sphaeroides 2.4.1],4P02_A Chain A, Cellulose Synthase subunit A [Cereibacter sphaeroides 2.4.1],5EIY_A Chain A, Putative cellulose synthase [Cereibacter sphaeroides 2.4.1],5EJZ_A Chain A, Putative cellulose synthase [Cereibacter sphaeroides 2.4.1] |
|
| 1.18e-14 | 280 | 790 | 237 | 564 | Chain A, Cellulose synthase catalytic subunit [UDP-forming] [Escherichia coli K-12] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 9.41e-24 | 452 | 711 | 584 | 764 | Cellulose synthase catalytic subunit A [UDP-forming] OS=Dictyostelium discoideum OX=44689 GN=dcsA PE=1 SV=1 |
|
| 6.01e-14 | 280 | 790 | 237 | 564 | Cellulose synthase catalytic subunit [UDP-forming] OS=Escherichia coli (strain K12) OX=83333 GN=bcsA PE=1 SV=3 |
|
| 7.15e-14 | 280 | 711 | 113 | 382 | Cellulose synthase catalytic subunit [UDP-forming] OS=Komagataeibacter sucrofermentans (strain ATCC 700178 / DSM 15973 / CECT 7291 / JCM 9730 / LMG 18788 / BPR 2001) OX=1307942 GN=bcsA PE=3 SV=1 |
|
| 7.90e-14 | 280 | 790 | 237 | 564 | Cellulose synthase catalytic subunit [UDP-forming] OS=Escherichia coli O157:H7 OX=83334 GN=bcsA PE=3 SV=2 |
|
| 1.80e-13 | 280 | 790 | 237 | 564 | Cellulose synthase catalytic subunit [UDP-forming] OS=Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) OX=99287 GN=bcsA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
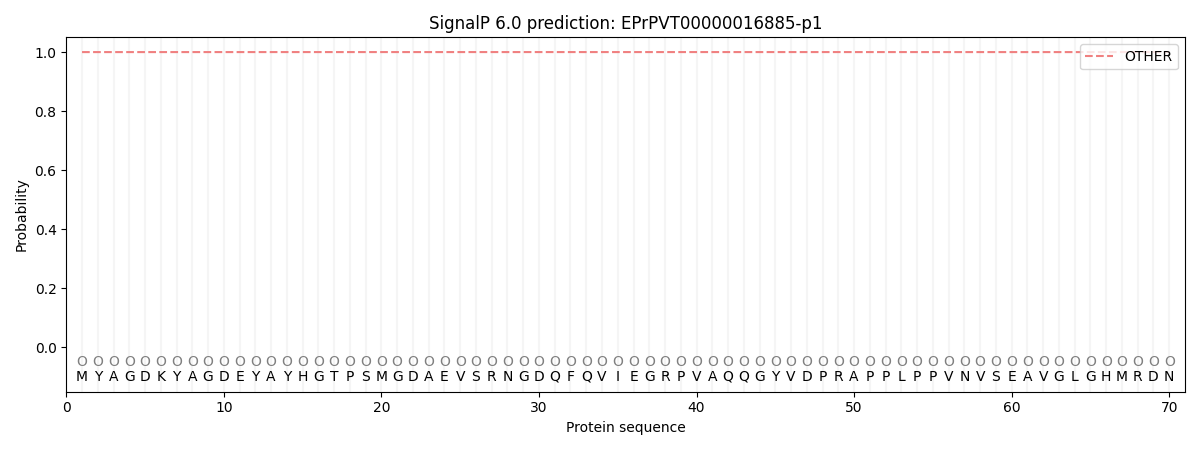
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000077 | 0.000001 |
TMHMM Annotations download full data without filtering help
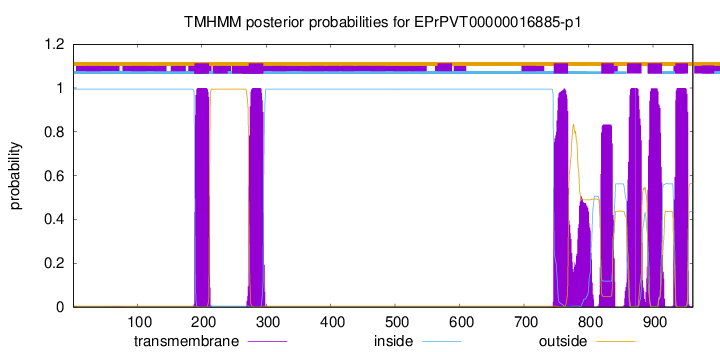
| Start | End |
|---|---|
| 189 | 211 |
| 273 | 295 |
| 746 | 768 |
| 820 | 839 |
| 860 | 882 |
| 892 | 914 |
| 935 | 954 |
