You are browsing environment: FUNGIDB
CAZyme Information: EPrPVT00000015633-p1
You are here: Home > Sequence: EPrPVT00000015633-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytopythium vexans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Pythiaceae; Phytopythium; Phytopythium vexans | |||||||||||
| CAZyme ID | EPrPVT00000015633-p1 | |||||||||||
| CAZy Family | CBM48 | |||||||||||
| CAZyme Description | Bifunctional purine biosynthesis protein PURH | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.21:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 112 | 324 | 1.3e-56 | 0.9351851851851852 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 223216 | PurH | 0.0 | 833 | 1400 | 1 | 515 | AICAR transformylase/IMP cyclohydrolase PurH [Nucleotide transport and metabolism]. |
| 180841 | PRK07106 | 0.0 | 1012 | 1400 | 3 | 390 | phosphoribosylaminoimidazolecarboxamide formyltransferase. |
| 234854 | purH | 1.75e-163 | 831 | 1400 | 1 | 513 | bifunctional phosphoribosylaminoimidazolecarboxamide formyltransferase/IMP cyclohydrolase; Provisional |
| 396396 | AICARFT_IMPCHas | 8.05e-99 | 938 | 1270 | 2 | 308 | AICARFT/IMPCHase bienzyme. This is a family of bifunctional enzymes catalyzing the last two steps in de novo purine biosynthesis. The bifunctional enzyme is found in both prokaryotes and eukaryotes. The second last step is catalyzed by 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase EC:2.1.2.3 (AICARFT), this enzyme catalyzes the formylation of AICAR with 10-formyl-tetrahydrofolate to yield FAICAR and tetrahydrofolate. This is catalyzed by a pair of C-terminal deaminase fold domains in the protein, where the active site is formed by the dimeric interface of two monomeric units. The last step is catalyzed by the N-terminal IMP (Inosine monophosphate) cyclohydrolase domain EC:3.5.4.10 (IMPCHase), cyclizing FAICAR (5-formylaminoimidazole-4-carboxamide ribonucleotide) to IMP. |
| 214822 | AICARFT_IMPCHas | 1.20e-96 | 937 | 1270 | 1 | 310 | AICARFT/IMPCHase bienzyme. This is a family of bifunctional enzymes catalysing the last two steps in de novo purine biosynthesis. The bifunctional enzyme is found in both prokaryotes and eukaryotes. The second last step is catalysed by 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase (AICARFT), this enzyme catalyses the formylation of AICAR with 10-formyl-tetrahydrofolate to yield FAICAR and tetrahydrofolate. The last step is catalysed by IMP (Inosine monophosphate) cyclohydrolase (IMPCHase), cyclizing FAICAR (5-formylaminoimidazole-4-carboxamide ribonucleotide) to IMP. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 9.05e-289 | 17 | 757 | 18 | 762 | |
| 4.29e-262 | 45 | 758 | 1 | 725 | |
| 1.96e-255 | 28 | 757 | 67 | 808 | |
| 8.43e-254 | 1 | 763 | 1 | 769 | |
| 2.86e-253 | 1 | 763 | 46 | 825 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.17e-156 | 830 | 1400 | 9 | 601 | Crystal structure of AICARFT bound to an antifolate [Homo sapiens],5UY8_B Crystal structure of AICARFT bound to an antifolate [Homo sapiens],5UY8_C Crystal structure of AICARFT bound to an antifolate [Homo sapiens],5UY8_D Crystal structure of AICARFT bound to an antifolate [Homo sapiens],5UZ0_A Crystal structure of AICARFT bound to an antifolate [Homo sapiens],5UZ0_B Crystal structure of AICARFT bound to an antifolate [Homo sapiens],5UZ0_C Crystal structure of AICARFT bound to an antifolate [Homo sapiens],5UZ0_D Crystal structure of AICARFT bound to an antifolate [Homo sapiens] |
|
| 6.56e-156 | 831 | 1400 | 1 | 592 | Crystal Structure of Human ATIC in complex with folate-based inhibitor BW1540U88UD [Homo sapiens],1P4R_B Crystal Structure of Human ATIC in complex with folate-based inhibitor BW1540U88UD [Homo sapiens],1PKX_A Crystal Structure of human ATIC in complex with XMP [Homo sapiens],1PKX_B Crystal Structure of human ATIC in complex with XMP [Homo sapiens],1PKX_C Crystal Structure of human ATIC in complex with XMP [Homo sapiens],1PKX_D Crystal Structure of human ATIC in complex with XMP [Homo sapiens],1PL0_A Crystal structure of human ATIC in complex with folate-based inhibitor, BW2315U89UC [Homo sapiens],1PL0_B Crystal structure of human ATIC in complex with folate-based inhibitor, BW2315U89UC [Homo sapiens],1PL0_C Crystal structure of human ATIC in complex with folate-based inhibitor, BW2315U89UC [Homo sapiens],1PL0_D Crystal structure of human ATIC in complex with folate-based inhibitor, BW2315U89UC [Homo sapiens] |
|
| 1.94e-154 | 848 | 1400 | 47 | 593 | Crystal Structure of Avian AICAR Transformylase in Complex with a Novel Inhibitor Identified by Virtual Ligand Screening [Gallus gallus],1THZ_B Crystal Structure of Avian AICAR Transformylase in Complex with a Novel Inhibitor Identified by Virtual Ligand Screening [Gallus gallus],2B1G_A Crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase [Gallus gallus],2B1G_B Crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase [Gallus gallus],2B1G_C Crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase [Gallus gallus],2B1G_D Crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase [Gallus gallus],2B1I_A crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase [Gallus gallus],2B1I_B crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase [Gallus gallus],2IU0_A crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase [Gallus gallus],2IU0_B crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase [Gallus gallus],2IU3_A Crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase [Gallus gallus],2IU3_B Crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase [Gallus gallus] |
|
| 3.68e-154 | 848 | 1400 | 67 | 613 | Crystal Structure Of The Homodimeric Bifunctional Transformylase And Cyclohydrolase Enzyme Avian Atic In Complex With Aicar And Xmp At 1.93 Angstroms. [Gallus gallus],1M9N_B Crystal Structure Of The Homodimeric Bifunctional Transformylase And Cyclohydrolase Enzyme Avian Atic In Complex With Aicar And Xmp At 1.93 Angstroms. [Gallus gallus],1OZ0_A Crystal Structure Of The Homodimeric Bifunctional Transformylase And Cyclohydrolase Enzyme Avian Atic In Complex With A Multisubstrate Adduct Inhibitor Beta-dadf. [Gallus gallus],1OZ0_B Crystal Structure Of The Homodimeric Bifunctional Transformylase And Cyclohydrolase Enzyme Avian Atic In Complex With A Multisubstrate Adduct Inhibitor Beta-dadf. [Gallus gallus] |
|
| 8.46e-152 | 837 | 1400 | 8 | 605 | Chain A, 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase [Cryptococcus neoformans],7MGQ_B Chain B, 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase [Cryptococcus neoformans],7MGQ_C Chain C, 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase [Cryptococcus neoformans],7MGQ_D Chain D, 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase [Cryptococcus neoformans] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.23e-155 | 831 | 1400 | 1 | 592 | Bifunctional purine biosynthesis protein ATIC OS=Pongo abelii OX=9601 GN=ATIC PE=2 SV=1 |
|
| 3.37e-155 | 831 | 1400 | 1 | 592 | Bifunctional purine biosynthesis protein ATIC OS=Homo sapiens OX=9606 GN=ATIC PE=1 SV=3 |
|
| 2.52e-154 | 831 | 1400 | 1 | 592 | Bifunctional purine biosynthesis protein ATIC OS=Bos taurus OX=9913 GN=ATIC PE=2 SV=1 |
|
| 9.65e-154 | 831 | 1400 | 1 | 592 | Bifunctional purine biosynthesis protein ATIC OS=Mus musculus OX=10090 GN=Atic PE=1 SV=2 |
|
| 9.96e-154 | 848 | 1400 | 47 | 593 | Bifunctional purine biosynthesis protein ATIC OS=Gallus gallus OX=9031 GN=ATIC PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
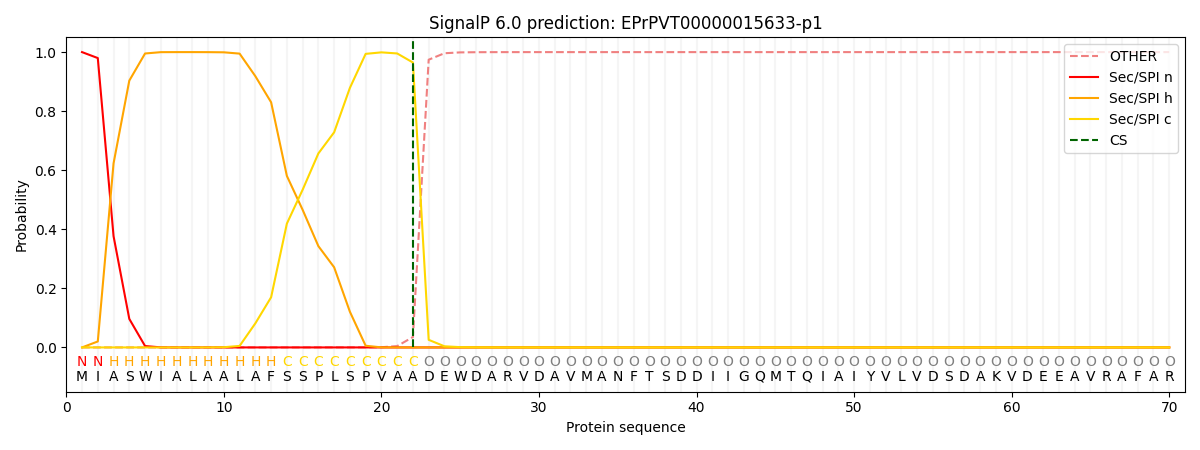
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000279 | 0.999691 | CS pos: 22-23. Pr: 0.9647 |
