You are browsing environment: FUNGIDB
CAZyme Information: EPrPVT00000014249-p1
Basic Information
help
| Species |
Phytopythium vexans
|
| Lineage |
Oomycota; NA; ; Pythiaceae; Phytopythium; Phytopythium vexans
|
| CAZyme ID |
EPrPVT00000014249-p1
|
| CAZy Family |
AA17|AA17 |
| CAZyme Description |
Glycoside hydrolase
|
| CAZyme Property |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_PvexansDAOMBR484 |
11991 |
1223560 |
34 |
11957
|
|
| Gene Location |
No EC number prediction in EPrPVT00000014249-p1.
| Family |
Start |
End |
Evalue |
family coverage |
| GH17 |
89 |
329 |
7e-20 |
0.887459807073955 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| 227625
|
Scw11 |
1.02e-20 |
53 |
327 |
47 |
303 |
Exo-beta-1,3-glucanase, GH17 family [Carbohydrate transport and metabolism]. |
| 366033
|
Glyco_hydro_17 |
3.84e-04 |
125 |
334 |
71 |
306 |
Glycosyl hydrolases family 17. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 3.00e-98 |
31 |
341 |
284 |
593 |
| 1.56e-92 |
46 |
339 |
87 |
381 |
| 4.56e-92 |
53 |
340 |
24 |
319 |
| 1.82e-91 |
55 |
325 |
27 |
301 |
| 4.09e-87 |
39 |
300 |
299 |
559 |
EPrPVT00000014249-p1 has no PDB hit.
EPrPVT00000014249-p1 has no Swissprot hit.
This protein is predicted as SP
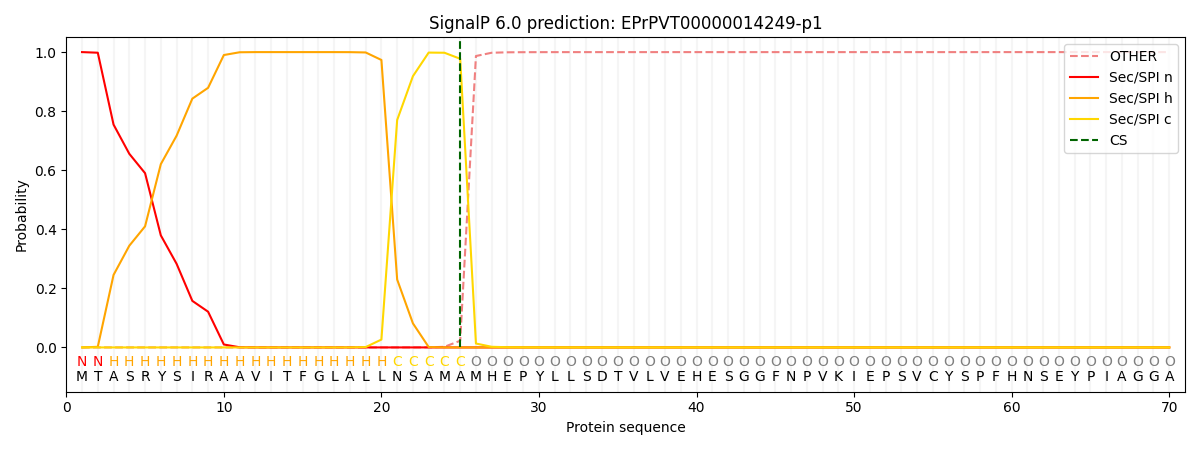
| Other |
SP_Sec_SPI |
CS Position |
| 0.000219 |
0.999754 |
CS pos: 25-26. Pr: 0.9774 |
There is no transmembrane helices in EPrPVT00000014249-p1.

