You are browsing environment: FUNGIDB
CAZyme Information: EPrPRT00000025171-p1
You are here: Home > Sequence: EPrPRT00000025171-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Pythium arrhenomanes | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Pythiaceae; Pythium; Pythium arrhenomanes | |||||||||||
| CAZyme ID | EPrPRT00000025171-p1 | |||||||||||
| CAZy Family | GT61 | |||||||||||
| CAZyme Description | Glucan 1,3-beta-glucosidase. | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 117 | 261 | 4.7e-78 | 0.5179856115107914 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 225344 | BglC | 2.50e-15 | 101 | 237 | 49 | 206 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 5.06e-117 | 50 | 261 | 63 | 311 | |
| 1.39e-101 | 50 | 261 | 42 | 289 | |
| 5.49e-101 | 48 | 261 | 38 | 287 | |
| 7.25e-97 | 47 | 261 | 16 | 267 | |
| 2.95e-96 | 47 | 261 | 81 | 332 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.40e-20 | 100 | 214 | 43 | 156 | Exo-b-(1,3)-glucanase From Candida Albicans [Candida albicans] |
|
| 8.44e-20 | 100 | 214 | 43 | 156 | Exo-b-(1,3)-glucanase From Candida Albicans At 1.85 A Resolution [Candida albicans],1EQC_A Exo-b-(1,3)-glucanase From Candida Albicans In Complex With Castanospermine At 1.85 A [Candida albicans] |
|
| 8.75e-20 | 100 | 214 | 48 | 161 | The structure of E292S glycosynthase variant of exo-1,3-beta-glucanase from Candida albicans at 1.85A resolution [Candida albicans SC5314],4M81_A The structure of E292S glycosynthase variant of exo-1,3-beta-glucanase from Candida albicans complexed with 1-fluoro-alpha-D-glucopyranoside (donor) and p-nitrophenyl beta-D-glucopyranoside (acceptor) at 1.86A resolution [Candida albicans SC5314],4M82_A The structure of E292S glycosynthase variant of exo-1,3-beta-glucanase from Candida albicans complexed with p-nitrophenyl-gentiobioside (product) at 1.6A resolution [Candida albicans SC5314] |
|
| 8.75e-20 | 100 | 214 | 48 | 161 | F229A/E292S Double Mutant of Exo-beta-1,3-glucanase from Candida albicans in Complex with Laminaritriose at 1.7 A [Candida albicans] |
|
| 8.80e-20 | 100 | 214 | 49 | 162 | Chain A, Hypothetical protein XOG1 [Candida albicans] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.80e-25 | 100 | 209 | 67 | 175 | Glucan 1,3-beta-glucosidase OS=Blumeria graminis OX=34373 PE=3 SV=1 |
|
| 1.29e-23 | 100 | 207 | 78 | 184 | Glucan 1,3-beta-glucosidase OS=Pichia angusta OX=870730 PE=3 SV=1 |
|
| 1.88e-23 | 99 | 207 | 58 | 165 | Probable glucan 1,3-beta-glucosidase A OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=exgA PE=3 SV=1 |
|
| 1.88e-23 | 99 | 207 | 58 | 165 | Glucan 1,3-beta-glucosidase A OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=exgA PE=1 SV=1 |
|
| 2.86e-23 | 99 | 207 | 69 | 176 | Probable glucan 1,3-beta-glucosidase A OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=exgA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
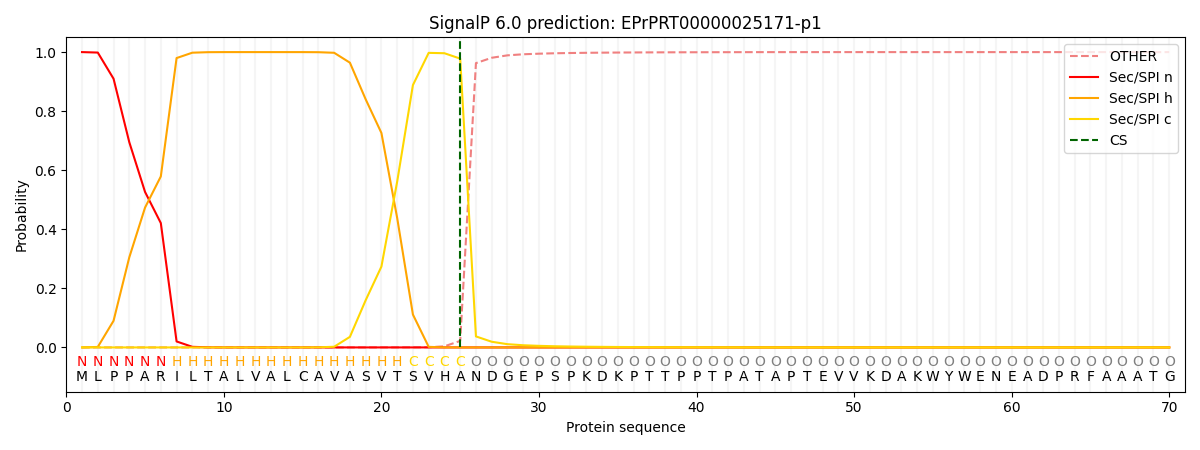
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000253 | 0.999733 | CS pos: 25-26. Pr: 0.9779 |
