You are browsing environment: FUNGIDB
CAZyme Information: EPrPRT00000022770-p1
You are here: Home > Sequence: EPrPRT00000022770-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Pythium arrhenomanes | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Pythiaceae; Pythium; Pythium arrhenomanes | |||||||||||
| CAZyme ID | EPrPRT00000022770-p1 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | Polysaccharide lyase. | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 4.2.2.2:4 | 2.4.1.186:3 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL3 | 36 | 223 | 1e-71 | 0.9639175257731959 |
| GT8 | 316 | 518 | 1.7e-32 | 0.8015564202334631 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397360 | Pectate_lyase | 5.48e-89 | 32 | 228 | 4 | 192 | Pectate lyase. |
| 133018 | GT8_Glycogenin | 3.21e-63 | 316 | 518 | 1 | 189 | Glycogenin belongs the GT 8 family and initiates the biosynthesis of glycogen. Glycogenin initiates the biosynthesis of glycogen by incorporating glucose residues through a self-glucosylation reaction at a Tyr residue, and then acts as substrate for chain elongation by glycogen synthase and branching enzyme. It contains a conserved DxD motif and an N-terminal beta-alpha-beta Rossmann-like fold that are common to the nucleotide-binding domains of most glycosyltransferases. The DxD motif is essential for coordination of the catalytic divalent cation, most commonly Mn2+. Glycogenin can be classified as a retaining glycosyltransferase, based on the relative anomeric stereochemistry of the substrate and product in the reaction catalyzed. It is placed in glycosyltransferase family 8 which includes lipopolysaccharide glucose and galactose transferases and galactinol synthases. |
| 395373 | Fe-ADH | 4.83e-37 | 531 | 830 | 26 | 346 | Iron-containing alcohol dehydrogenase. |
| 341486 | Fe-ADH-like | 3.62e-34 | 528 | 829 | 27 | 376 | iron-containing alcohol dehydrogenases (Fe-ADH)-like. This family contains iron-containing alcohol dehydrogenase (Fe-ADH) which catalyzes the reduction of acetaldehyde to alcohol with NADP as cofactor. Its activity requires iron ions. The protein structure represents a dehydroquinate synthase-like fold and is a member of the iron-activated alcohol dehydrogenase-like family. It is distinct from other alcohol dehydrogenases which contains different protein domains. Proteins of this family have not been characterized. |
| 341485 | Fe-ADH-like | 1.99e-32 | 568 | 723 | 68 | 222 | iron-containing alcohol dehydrogenases (Fe-ADH)-like. This family contains iron-containing alcohol dehydrogenase (Fe-ADH) which catalyzes the reduction of acetaldehyde to alcohol with NADP as cofactor. Its activity requires iron ions. The protein structure represents a dehydroquinate synthase-like fold and is a member of the iron-activated alcohol dehydrogenase-like family. It is distinct from other alcohol dehydrogenases which contains different protein domains. Proteins of this family have not been characterized. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.50e-83 | 23 | 255 | 27 | 257 | |
| 1.90e-82 | 24 | 254 | 18 | 248 | |
| 4.44e-81 | 23 | 255 | 27 | 257 | |
| 7.87e-80 | 316 | 530 | 2 | 217 | |
| 1.73e-64 | 25 | 249 | 21 | 266 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.13e-102 | 315 | 530 | 7 | 224 | UDP-galactose:glucoside-Skp1 alpha-D-galactosyltransferase with bound UDP and Platinum [Globisporangium ultimum],6MW8_A UDP-galactose:glucoside-Skp1 alpha-D-galactosyltransferase with bound UDP and Manganese [Globisporangium ultimum] |
|
| 1.80e-28 | 316 | 510 | 5 | 189 | Crystal Structure of Human Glycogenin-1 (GYG1), apo form [Homo sapiens],3QVB_A Crystal Structure of Human Glycogenin-1 (GYG1) complexed with manganese and UDP [Homo sapiens],3U2W_A Crystal Structure of Human Glycogenin-1 (GYG1) complexed with manganese and glucose or a glucal species [Homo sapiens],3U2W_B Crystal Structure of Human Glycogenin-1 (GYG1) complexed with manganese and glucose or a glucal species [Homo sapiens] |
|
| 1.80e-28 | 316 | 510 | 5 | 189 | Crystal Structure of Human Glycogenin-1 (GYG1) Tyr195pIPhe mutant, apo form [Homo sapiens],6EQL_A Crystal Structure of Human Glycogenin-1 (GYG1) Tyr195pIPhe mutant complexed with manganese and UDP [Homo sapiens],6EQL_B Crystal Structure of Human Glycogenin-1 (GYG1) Tyr195pIPhe mutant complexed with manganese and UDP [Homo sapiens] |
|
| 1.80e-28 | 316 | 510 | 5 | 189 | Crystal Structure of Human Glycogenin-1 (GYG1) complexed with manganese and UDP, in a triclinic closed form [Homo sapiens],3T7M_B Crystal Structure of Human Glycogenin-1 (GYG1) complexed with manganese and UDP, in a triclinic closed form [Homo sapiens],3T7N_A Crystal Structure of Human Glycogenin-1 (GYG1) complexed with manganese and UDP, in a monoclinic closed form [Homo sapiens],3T7N_B Crystal Structure of Human Glycogenin-1 (GYG1) complexed with manganese and UDP, in a monoclinic closed form [Homo sapiens],3T7O_A Crystal Structure of Human Glycogenin-1 (GYG1) complexed with manganese, UDP-Glucose and glucose [Homo sapiens],3T7O_B Crystal Structure of Human Glycogenin-1 (GYG1) complexed with manganese, UDP-Glucose and glucose [Homo sapiens],3U2U_A Crystal Structure of Human Glycogenin-1 (GYG1) complexed with manganese, UDP and maltotetraose [Homo sapiens],3U2U_B Crystal Structure of Human Glycogenin-1 (GYG1) complexed with manganese, UDP and maltotetraose [Homo sapiens],3U2V_A Crystal Structure of Human Glycogenin-1 (GYG1) complexed with manganese, UDP and maltohexaose [Homo sapiens],3U2V_B Crystal Structure of Human Glycogenin-1 (GYG1) complexed with manganese, UDP and maltohexaose [Homo sapiens],3U2X_A Crystal Structure of Human Glycogenin-1 (GYG1) complexed with manganese, UDP and 1'-deoxyglucose [Homo sapiens],3U2X_B Crystal Structure of Human Glycogenin-1 (GYG1) complexed with manganese, UDP and 1'-deoxyglucose [Homo sapiens] |
|
| 2.44e-28 | 316 | 510 | 5 | 189 | Crystal Structure of Human Glycogenin-1 (GYG1) T83M mutant complexed with manganese and UDP [Homo sapiens],3RMW_A Crystal Structure of Human Glycogenin-1 (GYG1) T83M mutant complexed with manganese and UDP-glucose [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.52e-50 | 33 | 213 | 43 | 220 | Probable pectate lyase D OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=plyD PE=3 SV=1 |
|
| 7.79e-48 | 33 | 244 | 41 | 247 | Pectate lyase H OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=plyH PE=1 SV=1 |
|
| 8.98e-48 | 33 | 244 | 37 | 241 | Probable pectate lyase E OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=plyE PE=3 SV=1 |
|
| 1.38e-47 | 33 | 244 | 28 | 233 | Probable pectate lyase G OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=plyG PE=3 SV=1 |
|
| 2.32e-47 | 33 | 244 | 37 | 241 | Probable pectate lyase E OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=plyE PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
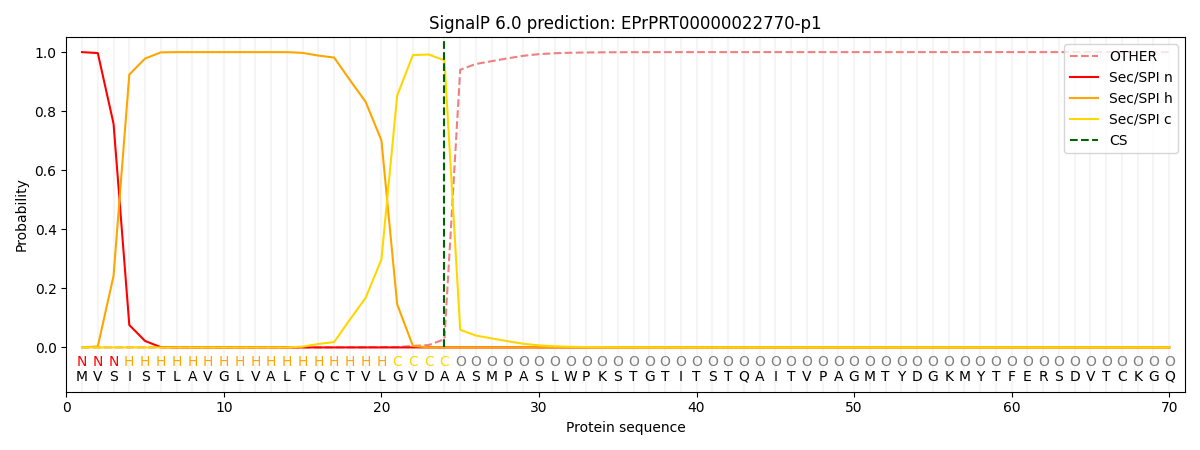
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000210 | 0.999743 | CS pos: 24-25. Pr: 0.9722 |
