You are browsing environment: FUNGIDB
CAZyme Information: EPrPRT00000021149-p1
You are here: Home > Sequence: EPrPRT00000021149-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
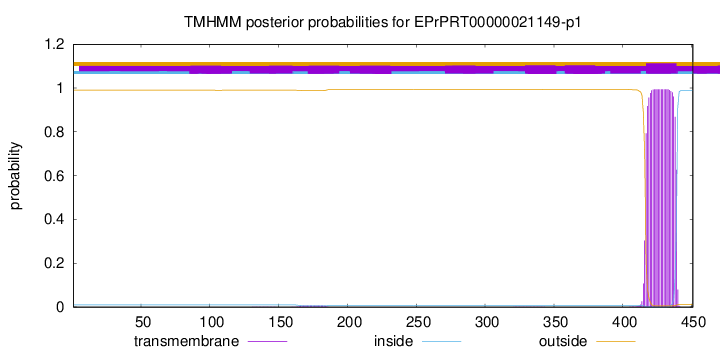
TMHMM annotations
Basic Information help
| Species | Pythium arrhenomanes | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Pythiaceae; Pythium; Pythium arrhenomanes | |||||||||||
| CAZyme ID | EPrPRT00000021149-p1 | |||||||||||
| CAZy Family | GH6 | |||||||||||
| CAZyme Description | Glycoside hydrolase. | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.21:2 | 3.2.1.23:1 | 3.2.1.38:1 | 3.2.1.-:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH1 | 2 | 395 | 1.7e-89 | 0.7785547785547785 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395176 | Glyco_hydro_1 | 1.09e-89 | 2 | 396 | 91 | 450 | Glycosyl hydrolase family 1. |
| 225343 | BglB | 8.42e-72 | 2 | 404 | 93 | 460 | Beta-glucosidase/6-phospho-beta-glucosidase/beta-galactosidase [Carbohydrate transport and metabolism]. |
| 215435 | PLN02814 | 4.18e-55 | 3 | 390 | 111 | 476 | beta-glucosidase |
| 184102 | PRK13511 | 5.44e-55 | 2 | 396 | 87 | 465 | 6-phospho-beta-galactosidase; Provisional |
| 215455 | PLN02849 | 6.18e-50 | 3 | 390 | 113 | 476 | beta-glucosidase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.48e-181 | 1 | 439 | 99 | 537 | |
| 4.44e-180 | 1 | 418 | 118 | 534 | |
| 1.49e-143 | 1 | 403 | 109 | 510 | |
| 5.39e-124 | 1 | 400 | 43 | 419 | |
| 3.54e-71 | 2 | 392 | 90 | 451 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.61e-60 | 2 | 401 | 99 | 467 | Chimeric Family 1 beta-glucosidase made with non-contiguous SCHEMA [Thermotoga maritima MSB8],4GXP_B Chimeric Family 1 beta-glucosidase made with non-contiguous SCHEMA [Thermotoga maritima MSB8],4GXP_C Chimeric Family 1 beta-glucosidase made with non-contiguous SCHEMA [Thermotoga maritima MSB8] |
|
| 8.04e-58 | 2 | 396 | 103 | 488 | Crystal Structure of a Rice Os3BGlu6 Beta-Glucosidase [Oryza sativa Japonica Group],3GNP_A Crystal Structure of a Rice Os3BGlu6 Beta-Glucosidase with Octyl-Beta-D-Thio-Glucoside [Oryza sativa Japonica Group],3GNR_A Crystal Structure of a Rice Os3BGlu6 Beta-Glucosidase with covalently bound 2-deoxy-2-fluoroglucoside to the catalytic nucleophile E396 [Oryza sativa Japonica Group] |
|
| 9.25e-58 | 3 | 396 | 94 | 461 | Aphid myrosinase [Brevicoryne brassicae],1WCG_B Aphid myrosinase [Brevicoryne brassicae] |
|
| 1.08e-57 | 3 | 365 | 118 | 465 | Chain A, Glycoside hydrolase [Nannochloris],5YJ7_B Chain B, Glycoside hydrolase [Nannochloris],5YJ7_C Chain C, Glycoside hydrolase [Nannochloris],5YJ7_D Chain D, Glycoside hydrolase [Nannochloris] |
|
| 2.24e-57 | 2 | 396 | 104 | 489 | Rice Os3BGlu6 E178Q with Covalent Glucosyl Moiety from p-nitrophenyl glucopyranoside. [Oryza sativa Japonica Group] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.37e-66 | 3 | 392 | 108 | 487 | Beta-glucosidase 27 OS=Arabidopsis thaliana OX=3702 GN=BGLU27 PE=2 SV=2 |
|
| 1.11e-61 | 3 | 392 | 120 | 500 | Beta-glucosidase 30 OS=Arabidopsis thaliana OX=3702 GN=BGLU30 PE=2 SV=1 |
|
| 4.82e-61 | 3 | 391 | 126 | 506 | Beta-glucosidase 32 OS=Arabidopsis thaliana OX=3702 GN=BGLU32 PE=2 SV=2 |
|
| 4.34e-60 | 3 | 392 | 109 | 490 | Beta-glucosidase 26, peroxisomal OS=Arabidopsis thaliana OX=3702 GN=BGLU26 PE=1 SV=1 |
|
| 1.85e-59 | 4 | 393 | 113 | 493 | Beta-glucosidase 25 OS=Oryza sativa subsp. japonica OX=39947 GN=BGLU25 PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000055 | 0.000000 |