You are browsing environment: FUNGIDB
CAZyme Information: EPrPRT00000020774-p1
You are here: Home > Sequence: EPrPRT00000020774-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Pythium arrhenomanes | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Pythiaceae; Pythium; Pythium arrhenomanes | |||||||||||
| CAZyme ID | EPrPRT00000020774-p1 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | Glycoside hydrolase. | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH6 | 46 | 305 | 5.2e-56 | 0.95578231292517 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396075 | Glyco_hydro_6 | 1.54e-40 | 54 | 303 | 2 | 287 | Glycosyl hydrolases family 6. |
| 227616 | CelA1 | 1.32e-17 | 89 | 371 | 178 | 463 | Cellulase/cellobiase CelA1 [Carbohydrate transport and metabolism]. |
| 411345 | gliding_GltJ | 1.51e-05 | 345 | 445 | 416 | 516 | adventurous gliding motility protein GltJ. Adventurous gliding motility protein GltJ, also known as AgmX, occurs in delta-proteobacteria such as Myxococcus xanthus. |
| 411345 | gliding_GltJ | 2.59e-05 | 345 | 447 | 408 | 512 | adventurous gliding motility protein GltJ. Adventurous gliding motility protein GltJ, also known as AgmX, occurs in delta-proteobacteria such as Myxococcus xanthus. |
| 223021 | PHA03247 | 2.95e-05 | 350 | 444 | 2662 | 2760 | large tegument protein UL36; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.08e-81 | 17 | 355 | 2 | 326 | |
| 2.44e-81 | 27 | 356 | 14 | 335 | |
| 3.47e-80 | 27 | 355 | 11 | 320 | |
| 4.34e-79 | 29 | 344 | 15 | 312 | |
| 1.01e-77 | 37 | 351 | 18 | 317 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.40e-29 | 41 | 331 | 16 | 317 | GH6 Orpinomyces sp. Y102 enzyme [Orpinomyces sp. Y102],5JX5_B GH6 Orpinomyces sp. Y102 enzyme [Orpinomyces sp. Y102],5JX5_C GH6 Orpinomyces sp. Y102 enzyme [Orpinomyces sp. Y102],5JX5_D GH6 Orpinomyces sp. Y102 enzyme [Orpinomyces sp. Y102],6IDW_A GH6 Orpinomyces sp. Y102 enzyme [Orpinomyces sp. Y102],6IDW_B GH6 Orpinomyces sp. Y102 enzyme [Orpinomyces sp. Y102],6IDW_C GH6 Orpinomyces sp. Y102 enzyme [Orpinomyces sp. Y102],6IDW_D GH6 Orpinomyces sp. Y102 enzyme [Orpinomyces sp. Y102] |
|
| 1.98e-28 | 52 | 347 | 28 | 357 | Crystal structure of a fungal chimeric cellobiohydrolase Cel6A [Humicola insolens] |
|
| 2.71e-28 | 99 | 331 | 73 | 291 | Structure of the endoglucanase Cel6 from Mycobacterium tuberculosis in complex with cellobiose at 1.75 angstrom [Mycobacterium tuberculosis H37Rv] |
|
| 2.71e-28 | 99 | 331 | 73 | 291 | Structure of the endoglucanase Cel6 from Mycobacterium tuberculosis in complex with METHYL-CELLOBIOSYL-4-DEOXY-4-THIO-BETA-D-CELLOBIOSIDE at 1.6 angstrom [Mycobacterium tuberculosis H37Rv] |
|
| 4.09e-28 | 99 | 331 | 94 | 312 | Structure of the endoglucanase Cel6 from Mycobacterium tuberculosis in complex with thiocellopentaose at 1.1 angstrom [Mycobacterium tuberculosis H37Rv] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.37e-25 | 71 | 301 | 75 | 301 | Endoglucanase A OS=Thermobispora bispora OX=2006 GN=celA PE=3 SV=1 |
|
| 7.40e-25 | 54 | 301 | 58 | 297 | Endoglucanase E-2 OS=Thermobifida fusca OX=2021 GN=celB PE=1 SV=2 |
|
| 2.09e-24 | 41 | 331 | 111 | 449 | Probable 1,4-beta-D-glucan cellobiohydrolase C OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=cbhC PE=3 SV=1 |
|
| 2.09e-24 | 41 | 331 | 111 | 449 | Probable 1,4-beta-D-glucan cellobiohydrolase C OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=cbhC PE=3 SV=1 |
|
| 3.66e-24 | 46 | 323 | 186 | 438 | Endoglucanase A OS=Cellulomonas fimi OX=1708 GN=cenA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
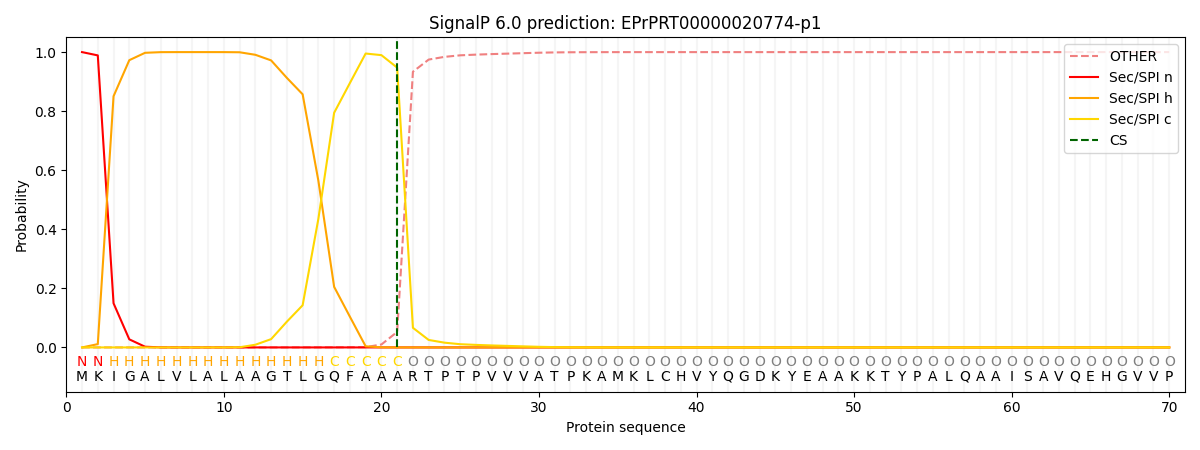
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000288 | 0.999689 | CS pos: 21-22. Pr: 0.9474 |
