You are browsing environment: FUNGIDB
CAZyme Information: EPrPRT00000020257-p1
You are here: Home > Sequence: EPrPRT00000020257-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Pythium arrhenomanes | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Pythiaceae; Pythium; Pythium arrhenomanes | |||||||||||
| CAZyme ID | EPrPRT00000020257-p1 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | Dolichyl-P-Man:Man(5)GlcNAc(2)-PP-dolichyl mannosyltransferase. | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.258:18 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT58 | 49 | 402 | 4.5e-136 | 0.9945054945054945 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 398745 | ALG3 | 0.0 | 51 | 402 | 3 | 358 | ALG3 protein. The formation of N-glycosidic linkages of glycoproteins involves the ordered assembly of the common Glc3Man9GlcNAc2 core-oligosaccharide on the lipid carrier dolichyl pyrophosphate. Whereas early mannosylation steps occur on the cytoplasmic side of the endoplasmic reticulum with GDP-Man as donor, the final reactions from Man5GlcNAc2-PP-Dol to Man9GlcNAc2-PP-Dol on the lumenal side use Dol-P-Man. ALG3 gene encodes the Dol-P-Man:Man5GlcNAc2-PP-Dol mannosyltransferase. |
| 273013 | diphth2_R | 3.87e-91 | 514 | 787 | 17 | 318 | diphthamide biosynthesis enzyme Dph1/Dph2 domain. Archaea and Eukaryotes, but not Eubacteria, share the property of having a covalently modified residue, 2'-[3-carboxamido-3-(trimethylammonio)propyl]histidine, as a part of a cytosolic protein. The modified His, termed diphthamide, is part of translation elongation factor EF-2 and is the site for ADP-ribosylation by diphtheria toxin. This model includes both Dph1 and Dph2 from Saccharomyces cerevisiae, although only Dph2 is found in the Archaea (see TIGR03682). Dph2 has been shown to act analogously to the radical SAM (rSAM) family (pfam04055), with 4Fe-4S-assisted cleavage of S-adenosylmethionine to create a free radical, but a different organic radical than in rSAM. |
| 272990 | DPH2 | 4.03e-87 | 514 | 925 | 51 | 496 | diphthamide biosynthesis protein 2. This protein has been shown in Saccharomyces cerevisiae to be one of several required for the modification of a particular histidine residue of translation elongation factor 2 to diphthamide. This modified site can then become the target for ADP-ribosylation by diphtheria toxin. [Protein fate, Protein modification and repair] |
| 396438 | Diphthamide_syn | 4.29e-87 | 521 | 795 | 1 | 299 | Putative diphthamide synthesis protein. Diphthamide_syn, diphthamide synthase, catalyzes the last amidation step of diphthamide biosynthesis using ammonium and ATP. Human DPH1 is a candidate tumor suppressor gene. DPH2 from yeast, which confers resistance to diphtheria toxin has been found to be involved in diphthamide synthesis. Diphtheria toxin inhibits eukaryotic protein synthesis by ADP-ribosylating diphthamide, a post-translationally modified histidine residue present in EF2. Diphthamide synthase is evolutionarily conserved in eukaryotes. Diphthamide is a post-translationally modified histidine residue found on archaeal and eukaryotic translation elongation factor 2 (eEF-2). |
| 224650 | DPH2 | 1.02e-49 | 505 | 820 | 19 | 345 | Diphthamide synthase subunit DPH2 [Translation, ribosomal structure and biogenesis]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.16e-231 | 13 | 508 | 12 | 560 | |
| 2.21e-216 | 12 | 494 | 9 | 539 | |
| 1.19e-101 | 35 | 407 | 27 | 405 | |
| 1.19e-101 | 35 | 407 | 27 | 405 | |
| 7.80e-101 | 48 | 407 | 49 | 403 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.22e-19 | 517 | 797 | 26 | 327 | Crystal structure of Methanobrevibacter smithii Dph2 in complex with Methanobrevibacter smithii elongation factor 2 [Methanobrevibacter smithii],6Q2D_B Crystal structure of Methanobrevibacter smithii Dph2 in complex with Methanobrevibacter smithii elongation factor 2 [Methanobrevibacter smithii],6Q2E_A Crystal structure of Methanobrevibacter smithii Dph2 bound to 5'-methylthioadenosine [Methanobrevibacter smithii],6Q2E_B Crystal structure of Methanobrevibacter smithii Dph2 bound to 5'-methylthioadenosine [Methanobrevibacter smithii] |
|
| 1.33e-12 | 517 | 805 | 58 | 360 | Crystal structure of Dph2 from Pyrococcus horikoshii [Pyrococcus horikoshii],3LZC_B Crystal structure of Dph2 from Pyrococcus horikoshii [Pyrococcus horikoshii],3LZD_A Crystal structure of Dph2 from Pyrococcus horikoshii with 4Fe-4S cluster [Pyrococcus horikoshii],3LZD_B Crystal structure of Dph2 from Pyrococcus horikoshii with 4Fe-4S cluster [Pyrococcus horikoshii],6BXK_A Crystal structure of Pyrococcus horikoshii Dph2 with 4Fe-4S cluster and MTA [Pyrococcus horikoshii OT3],6BXK_B Crystal structure of Pyrococcus horikoshii Dph2 with 4Fe-4S cluster and MTA [Pyrococcus horikoshii OT3],6BXL_A Crystal structure of Pyrococcus horikoshii Dph2 with 4Fe-4S cluster and SAM [Pyrococcus horikoshii OT3],6BXL_B Crystal structure of Pyrococcus horikoshii Dph2 with 4Fe-4S cluster and SAM [Pyrococcus horikoshii OT3] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.40e-95 | 48 | 409 | 22 | 414 | Probable Dol-P-Man:Man(5)GlcNAc(2)-PP-Dol alpha-1,3-mannosyltransferase OS=Dictyostelium discoideum OX=44689 GN=alg3 PE=3 SV=1 |
|
| 4.15e-95 | 35 | 410 | 33 | 414 | Dol-P-Man:Man(5)GlcNAc(2)-PP-Dol alpha-1,3-mannosyltransferase OS=Mus musculus OX=10090 GN=Alg3 PE=1 SV=2 |
|
| 3.11e-93 | 42 | 410 | 40 | 414 | Dol-P-Man:Man(5)GlcNAc(2)-PP-Dol alpha-1,3-mannosyltransferase OS=Homo sapiens OX=9606 GN=ALG3 PE=1 SV=1 |
|
| 2.31e-91 | 47 | 405 | 35 | 404 | Dol-P-Man:Man(5)GlcNAc(2)-PP-Dol alpha-1,3-mannosyltransferase OS=Arabidopsis thaliana OX=3702 GN=ALG3 PE=1 SV=1 |
|
| 1.40e-85 | 51 | 405 | 40 | 407 | Dol-P-Man:Man(5)GlcNAc(2)-PP-Dol alpha-1,3-mannosyltransferase OS=Neurospora crassa (strain ATCC 24698 / 74-OR23-1A / CBS 708.71 / DSM 1257 / FGSC 987) OX=367110 GN=alg-3 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999994 | 0.000048 |
TMHMM Annotations download full data without filtering help
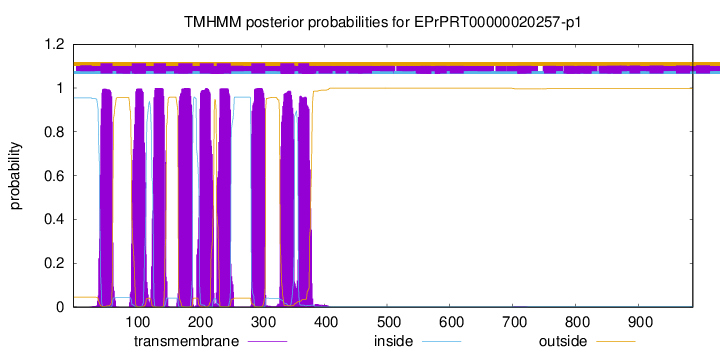
| Start | End |
|---|---|
| 44 | 63 |
| 94 | 116 |
| 129 | 148 |
| 168 | 190 |
| 202 | 224 |
| 229 | 251 |
| 284 | 306 |
| 330 | 352 |
| 359 | 376 |
