You are browsing environment: FUNGIDB
CAZyme Information: EPrPRT00000019341-p1
You are here: Home > Sequence: EPrPRT00000019341-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Pythium arrhenomanes | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Pythiaceae; Pythium; Pythium arrhenomanes | |||||||||||
| CAZyme ID | EPrPRT00000019341-p1 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | Callose synthase. | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.34:28 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT48 | 1 | 301 | 4.7e-66 | 0.35182679296346414 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396784 | Glucan_synthase | 6.63e-21 | 5 | 292 | 472 | 717 | 1,3-beta-glucan synthase component. This family consists of various 1,3-beta-glucan synthase components including Gls1, Gls2 and Gls3 from yeast. 1,3-beta-glucan synthase EC:2.4.1.34 also known as callose synthase catalyzes the formation of a beta-1,3-glucan polymer that is a major component of the fungal cell wall. The reaction catalyzed is:- UDP-glucose + {(1,3)-beta-D-glucosyl}(N) <=> UDP + {(1,3)-beta-D-glucosyl}(N+1). |
| 395036 | Sugar_tr | 9.45e-06 | 490 | 881 | 2 | 434 | Sugar (and other) transporter. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.42e-301 | 1 | 906 | 1291 | 2235 | |
| 2.18e-227 | 1 | 881 | 1307 | 2213 | |
| 6.12e-214 | 1 | 890 | 1292 | 2247 | |
| 1.05e-212 | 1 | 890 | 1267 | 2205 | |
| 9.49e-78 | 1 | 848 | 1273 | 2122 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.00e-38 | 1 | 486 | 1483 | 1922 | Callose synthase 7 OS=Arabidopsis thaliana OX=3702 GN=CALS7 PE=3 SV=3 |
|
| 3.73e-38 | 1 | 486 | 1326 | 1763 | Callose synthase 11 OS=Arabidopsis thaliana OX=3702 GN=CALS11 PE=2 SV=1 |
|
| 6.78e-38 | 1 | 479 | 1473 | 1910 | Callose synthase 5 OS=Arabidopsis thaliana OX=3702 GN=CALS5 PE=1 SV=1 |
|
| 1.12e-37 | 1 | 479 | 1333 | 1765 | Callose synthase 12 OS=Arabidopsis thaliana OX=3702 GN=CALS12 PE=2 SV=1 |
|
| 3.50e-37 | 6 | 488 | 1465 | 1898 | Callose synthase 10 OS=Arabidopsis thaliana OX=3702 GN=CALS10 PE=2 SV=5 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000062 | 0.000000 |
TMHMM Annotations download full data without filtering help
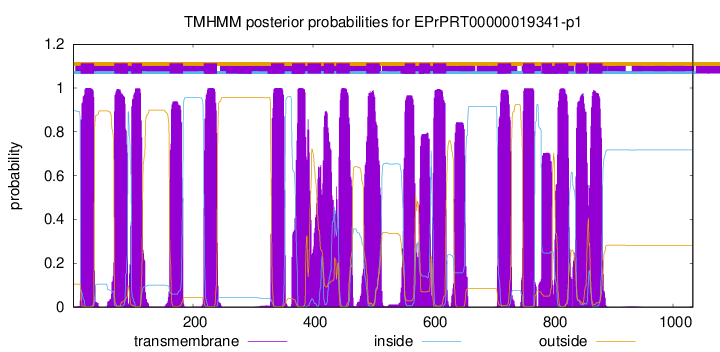
| Start | End |
|---|---|
| 13 | 35 |
| 69 | 91 |
| 98 | 115 |
| 161 | 183 |
| 218 | 240 |
| 330 | 352 |
| 365 | 387 |
| 391 | 413 |
| 418 | 435 |
| 439 | 461 |
| 489 | 511 |
| 553 | 570 |
| 577 | 595 |
| 600 | 622 |
| 708 | 730 |
| 750 | 769 |
| 781 | 798 |
| 803 | 825 |
| 838 | 856 |
| 860 | 882 |
