You are browsing environment: FUNGIDB
CAZyme Information: EPrPRT00000014722-p1
You are here: Home > Sequence: EPrPRT00000014722-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Pythium arrhenomanes | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Pythiaceae; Pythium; Pythium arrhenomanes | |||||||||||
| CAZyme ID | EPrPRT00000014722-p1 | |||||||||||
| CAZy Family | AA2 | |||||||||||
| CAZyme Description | T-complex protein 1 subunit eta. | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH81 | 305 | 562 | 1e-36 | 0.35691318327974275 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 239456 | TCP1_eta | 0.0 | 864 | 1380 | 1 | 516 | TCP-1 (CTT or eukaryotic type II) chaperonin family, eta subunit. Chaperonins are involved in productive folding of proteins. They share a common general morphology, a double toroid of 2 stacked rings. In contrast to bacterial group I chaperonins (GroEL), each ring of the eukaryotic cytosolic chaperonin (CTT) consists of eight different, but homologous subunits. Their common function is to sequester nonnative proteins inside their central cavity and promote folding by using energy derived from ATP hydrolysis. The best studied in vivo substrates of CTT are actin and tubulin. |
| 238189 | chaperonin_type_I_II | 0.0 | 872 | 1380 | 1 | 462 | chaperonin families, type I and type II. Chaperonins are involved in productive folding of proteins. They share a common general morphology, a double toroid of 2 stacked rings, each composed of 7-9 subunits. There are 2 main chaperonin groups. The symmetry of type I is seven-fold and they are found in eubacteria (GroEL) and in organelles of eubacterial descent (hsp60 and RBP). The symmetry of type II is eight- or nine-fold and they are found in archea (thermosome), thermophilic bacteria (TF55) and in the eukaryotic cytosol (CTT). Their common function is to sequester nonnative proteins inside their central cavity and promote folding by using energy derived from ATP hydrolysis. |
| 274086 | chap_CCT_eta | 0.0 | 862 | 1380 | 1 | 517 | T-complex protein 1, eta subunit. Members of this family, all eukaryotic, are part of the group II chaperonin complex called CCT (chaperonin containing TCP-1) or TRiC. The archaeal equivalent group II chaperonin is often called the thermosome. Both are somewhat related to the group I chaperonin of bacterial, GroEL/GroES. This family consists exclusively of the CCT eta chain (part of a paralogous family) from animals, plants, fungi, and other eukaryotes. |
| 395068 | Cpn60_TCP1 | 0.0 | 891 | 1379 | 1 | 484 | TCP-1/cpn60 chaperonin family. This family includes members from the HSP60 chaperone family and the TCP-1 (T-complex protein) family. |
| 239459 | cpn60 | 7.61e-160 | 865 | 1379 | 1 | 512 | cpn60 chaperonin family. Chaperonins are involved in productive folding of proteins. They share a common general morphology, a double toroid of 2 stacked rings. Archaeal cpn60 (thermosome), together with TF55 from thermophilic bacteria and the eukaryotic cytosol chaperonin (CTT), belong to the type II group of chaperonins. Cpn60 consists of two stacked octameric rings, which are composed of one or two different subunits. Their common function is to sequester nonnative proteins inside their central cavity and promote folding by using energy derived from ATP hydrolysis. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.43e-253 | 52 | 859 | 94 | 793 | |
| 1.43e-253 | 52 | 859 | 94 | 793 | |
| 4.01e-253 | 52 | 859 | 94 | 793 | |
| 4.01e-253 | 52 | 859 | 94 | 793 | |
| 1.94e-197 | 52 | 859 | 38 | 737 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.97e-230 | 865 | 1380 | 6 | 519 | Chain H, T-complex protein 1 subunit eta [Homo sapiens],6QB8_h Chain h, T-complex protein 1 subunit eta [Homo sapiens],7LUM_C Chain C, T-complex protein 1 subunit eta [Homo sapiens],7LUM_K Chain K, T-complex protein 1 subunit eta [Homo sapiens],7LUP_C Chain C, T-complex protein 1 subunit eta [Homo sapiens],7LUP_K Chain K, T-complex protein 1 subunit eta [Homo sapiens],7NVL_H Chain H, T-complex protein 1 subunit eta [Homo sapiens],7NVL_h Chain h, T-complex protein 1 subunit eta [Homo sapiens],7NVM_H Chain H, T-complex protein 1 subunit eta [Homo sapiens],7NVM_h Chain h, T-complex protein 1 subunit eta [Homo sapiens],7NVN_H Chain H, T-complex protein 1 subunit eta [Homo sapiens],7NVN_h Chain h, T-complex protein 1 subunit eta [Homo sapiens],7NVO_H Chain H, T-complex protein 1 subunit eta [Homo sapiens],7NVO_h Chain h, T-complex protein 1 subunit eta [Homo sapiens] |
|
| 7.01e-230 | 865 | 1380 | 6 | 519 | The crystal structures of the eukaryotic chaperonin CCT reveal its functional partitioning [Bos taurus],4B2T_h The crystal structures of the eukaryotic chaperonin CCT reveal its functional partitioning [Bos taurus] |
|
| 4.42e-228 | 869 | 1380 | 1 | 510 | Ca model of bovine TRiC/CCT derived from a 4.0 Angstrom cryo-EM map [Bos taurus] |
|
| 9.37e-227 | 871 | 1380 | 1 | 508 | hTRiC-hPFD Class6 [Homo sapiens],6NR8_O hTRiC-hPFD Class6 [Homo sapiens],6NR9_G hTRiC-hPFD Class5 [Homo sapiens],6NR9_O hTRiC-hPFD Class5 [Homo sapiens],6NRA_G hTRiC-hPFD Class1 (No PFD) [Homo sapiens],6NRA_O hTRiC-hPFD Class1 (No PFD) [Homo sapiens],6NRB_G hTRiC-hPFD Class2 [Homo sapiens],6NRB_O hTRiC-hPFD Class2 [Homo sapiens],6NRC_G hTRiC-hPFD Class3 [Homo sapiens],6NRC_O hTRiC-hPFD Class3 [Homo sapiens],6NRD_G hTRiC-hPFD Class4 [Homo sapiens],6NRD_O hTRiC-hPFD Class4 [Homo sapiens] |
|
| 3.45e-208 | 860 | 1380 | 4 | 523 | The crystal structure of yeast CCT reveals intrinsic asymmetry of eukaryotic cytosolic chaperonins [Saccharomyces cerevisiae],4V81_O The crystal structure of yeast CCT reveals intrinsic asymmetry of eukaryotic cytosolic chaperonins [Saccharomyces cerevisiae],4V81_g The crystal structure of yeast CCT reveals intrinsic asymmetry of eukaryotic cytosolic chaperonins [Saccharomyces cerevisiae],4V81_o The crystal structure of yeast CCT reveals intrinsic asymmetry of eukaryotic cytosolic chaperonins [Saccharomyces cerevisiae],4V8R_AH Chain AH, T-COMPLEX PROTEIN 1 SUBUNIT ETA [Saccharomyces cerevisiae],4V8R_Ah Chain Ah, T-COMPLEX PROTEIN 1 SUBUNIT ETA [Saccharomyces cerevisiae],4V8R_BH Chain BH, T-COMPLEX PROTEIN 1 SUBUNIT ETA [Saccharomyces cerevisiae],4V8R_Bh Chain Bh, T-COMPLEX PROTEIN 1 SUBUNIT ETA [Saccharomyces cerevisiae],4V94_G Molecular architecture of the eukaryotic chaperonin TRiC/CCT derived by a combination of chemical crosslinking and mass-spectrometry, XL-MS [Saccharomyces cerevisiae S288C],4V94_O Molecular architecture of the eukaryotic chaperonin TRiC/CCT derived by a combination of chemical crosslinking and mass-spectrometry, XL-MS [Saccharomyces cerevisiae S288C],4V94_g Molecular architecture of the eukaryotic chaperonin TRiC/CCT derived by a combination of chemical crosslinking and mass-spectrometry, XL-MS [Saccharomyces cerevisiae S288C],4V94_o Molecular architecture of the eukaryotic chaperonin TRiC/CCT derived by a combination of chemical crosslinking and mass-spectrometry, XL-MS [Saccharomyces cerevisiae S288C],5GW4_H Structure of Yeast NPP-TRiC [Saccharomyces cerevisiae S288C],5GW4_h Structure of Yeast NPP-TRiC [Saccharomyces cerevisiae S288C],5GW5_H Structure of TRiC-AMP-PNP [Saccharomyces cerevisiae S288C],5GW5_h Structure of TRiC-AMP-PNP [Saccharomyces cerevisiae S288C],6KRD_H TRiC at 0.05 mM ADP-AlFx, Conformation 4, 0.05-C4 [Saccharomyces cerevisiae S288C],6KRD_h TRiC at 0.05 mM ADP-AlFx, Conformation 4, 0.05-C4 [Saccharomyces cerevisiae S288C],6KRE_H TRiC at 0.05 mM ADP-AlFx, Conformation 2, 0.05-C2 [Saccharomyces cerevisiae S288C],6KRE_h TRiC at 0.05 mM ADP-AlFx, Conformation 2, 0.05-C2 [Saccharomyces cerevisiae S288C],6KS6_H TRiC at 0.2 mM ADP-AlFx, Conformation 1, 0.2-C1 [Saccharomyces cerevisiae S288C],6KS6_h TRiC at 0.2 mM ADP-AlFx, Conformation 1, 0.2-C1 [Saccharomyces cerevisiae S288C],6KS7_H TRiC at 0.1 mM ADP-AlFx, Conformation 1, 0.1-C1 [Saccharomyces cerevisiae S288C],6KS7_h TRiC at 0.1 mM ADP-AlFx, Conformation 1, 0.1-C1 [Saccharomyces cerevisiae S288C],6KS8_H TRiC at 0.1 mM ADP-AlFx, Conformation 4, 0.1-C4 [Saccharomyces cerevisiae S288C],6KS8_h TRiC at 0.1 mM ADP-AlFx, Conformation 4, 0.1-C4 [Saccharomyces cerevisiae S288C] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.90e-244 | 862 | 1380 | 6 | 525 | T-complex protein 1 subunit eta OS=Arabidopsis thaliana OX=3702 GN=CCT7 PE=1 SV=1 |
|
| 6.71e-230 | 865 | 1380 | 6 | 519 | T-complex protein 1 subunit eta OS=Mus musculus OX=10090 GN=Cct7 PE=1 SV=1 |
|
| 2.56e-229 | 865 | 1380 | 6 | 519 | T-complex protein 1 subunit eta OS=Homo sapiens OX=9606 GN=CCT7 PE=1 SV=2 |
|
| 3.60e-229 | 865 | 1380 | 6 | 519 | T-complex protein 1 subunit eta OS=Bos taurus OX=9913 GN=CCT7 PE=1 SV=1 |
|
| 3.60e-229 | 865 | 1380 | 6 | 519 | T-complex protein 1 subunit eta OS=Pongo abelii OX=9601 GN=CCT7 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
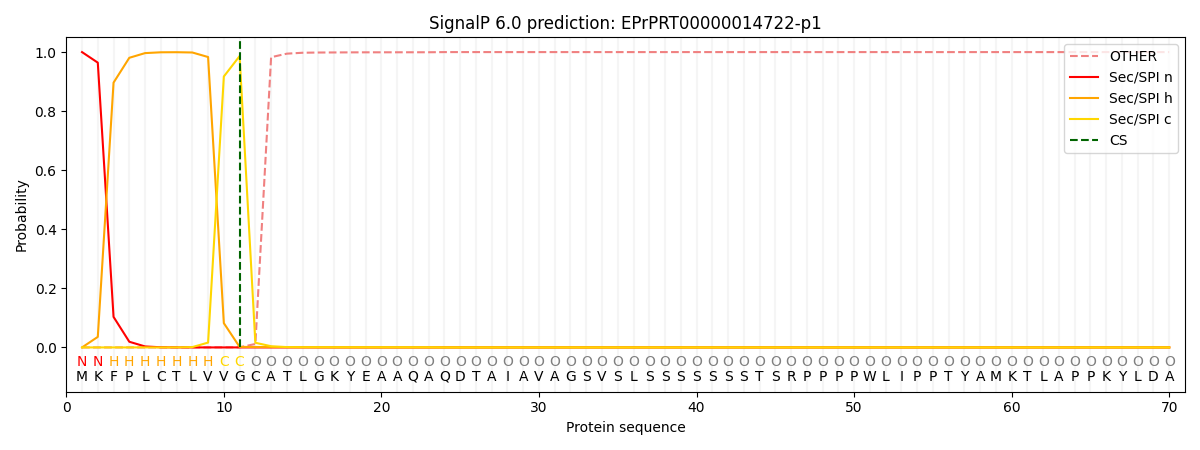
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000006 | 1.000018 | CS pos: 11-12. Pr: 0.9865 |
