You are browsing environment: FUNGIDB
CAZyme Information: EPrPRT00000014333-p1
You are here: Home > Sequence: EPrPRT00000014333-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Pythium arrhenomanes | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Pythiaceae; Pythium; Pythium arrhenomanes | |||||||||||
| CAZyme ID | EPrPRT00000014333-p1 | |||||||||||
| CAZy Family | AA17 | |||||||||||
| CAZyme Description | Beta-mannosidase. | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.25:5 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 26 | 412 | 9.3e-35 | 0.3603723404255319 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 225789 | LacZ | 2.37e-35 | 35 | 513 | 12 | 382 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| 407625 | Ig_mannosidase | 4.51e-08 | 751 | 817 | 9 | 74 | Ig-fold domain. This Ig-like fold domain is found in mannosidase enzymes. |
| 395572 | Glyco_hydro_2 | 1.89e-06 | 229 | 348 | 1 | 106 | Glycosyl hydrolases family 2. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
| 236673 | ebgA | 1.44e-04 | 298 | 494 | 272 | 397 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.23e-148 | 30 | 1027 | 62 | 988 | |
| 4.31e-141 | 25 | 1030 | 23 | 862 | |
| 2.03e-89 | 31 | 931 | 37 | 743 | |
| 4.17e-87 | 50 | 975 | 41 | 732 | |
| 1.07e-83 | 37 | 924 | 38 | 737 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.64e-76 | 32 | 925 | 17 | 707 | mouse beta-mannosidase (MANBA) [Mus musculus],6DDU_A mouse beta-mannosidase bound to beta-D-mannose (MANBA) [Mus musculus] |
|
| 6.13e-54 | 33 | 618 | 33 | 538 | Crystal structure of Beta-D-Mannosidase from Dictyoglomus thermophilum. [Dictyoglomus thermophilum H-6-12],5N6U_B Crystal structure of Beta-D-Mannosidase from Dictyoglomus thermophilum. [Dictyoglomus thermophilum H-6-12],5N6U_C Crystal structure of Beta-D-Mannosidase from Dictyoglomus thermophilum. [Dictyoglomus thermophilum H-6-12],5N6U_D Crystal structure of Beta-D-Mannosidase from Dictyoglomus thermophilum. [Dictyoglomus thermophilum H-6-12] |
|
| 1.37e-50 | 23 | 916 | 28 | 761 | Structure of Fungal beta-mannosidase from Glycoside Hydrolase Family 2 of Trichoderma harzianum [Trichoderma harzianum],4UOJ_A Chain A, BETA-MANNOSIDASE GH2 [Trichoderma harzianum],4UOJ_B Chain B, BETA-MANNOSIDASE GH2 [Trichoderma harzianum] |
|
| 4.45e-47 | 47 | 957 | 33 | 730 | Crystal structure of the acid-base mutant (E477A) of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri [Xanthomonas citri pv. citri str. 306],6BYI_B Crystal structure of the acid-base mutant (E477A) of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri [Xanthomonas citri pv. citri str. 306] |
|
| 4.48e-47 | 47 | 957 | 35 | 732 | Crystal structure of the nucleophile mutant (E575A) of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri [Xanthomonas citri pv. citri str. 306],6BYG_B Crystal structure of the nucleophile mutant (E575A) of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri [Xanthomonas citri pv. citri str. 306] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 9.18e-81 | 5 | 916 | 3 | 710 | Beta-mannosidase OS=Capra hircus OX=9925 GN=MANBA PE=1 SV=1 |
|
| 3.60e-78 | 5 | 897 | 3 | 692 | Beta-mannosidase OS=Bos taurus OX=9913 GN=MANBA PE=1 SV=1 |
|
| 6.21e-77 | 32 | 916 | 28 | 710 | Beta-mannosidase OS=Rattus norvegicus OX=10116 GN=Manba PE=2 SV=1 |
|
| 9.99e-76 | 32 | 925 | 28 | 718 | Beta-mannosidase OS=Mus musculus OX=10090 GN=Manba PE=1 SV=1 |
|
| 8.83e-75 | 5 | 916 | 3 | 710 | Beta-mannosidase OS=Homo sapiens OX=9606 GN=MANBA PE=1 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as SP
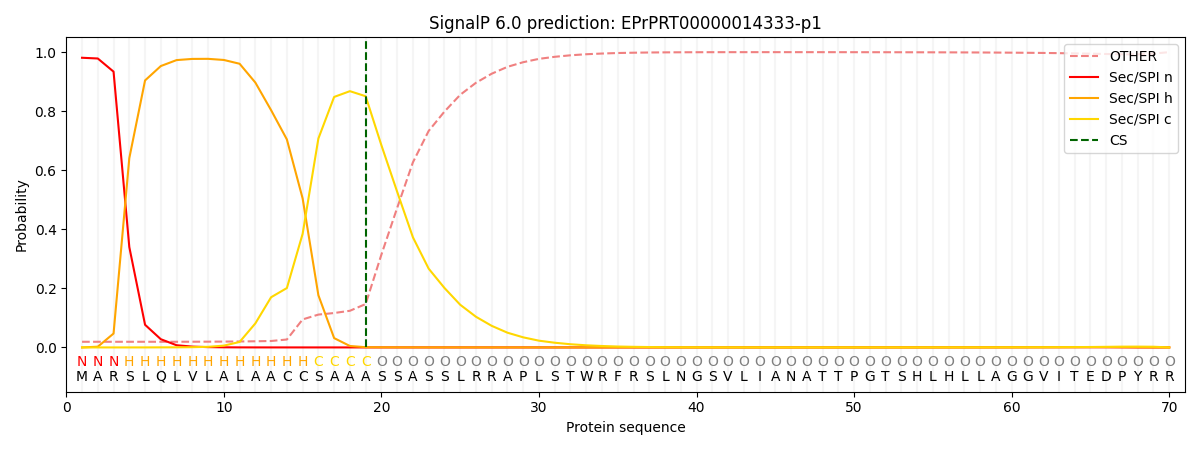
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.031969 | 0.967999 | CS pos: 19-20. Pr: 0.8506 |
