You are browsing environment: FUNGIDB
CAZyme Information: EPrPRT00000013465-p1
You are here: Home > Sequence: EPrPRT00000013465-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Pythium arrhenomanes | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Pythiaceae; Pythium; Pythium arrhenomanes | |||||||||||
| CAZyme ID | EPrPRT00000013465-p1 | |||||||||||
| CAZy Family | AA17 | |||||||||||
| CAZyme Description | Callose synthase. | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.34:28 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT48 | 453 | 1185 | 4.2e-257 | 0.9350473612990527 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396784 | Glucan_synthase | 3.63e-176 | 458 | 1138 | 7 | 700 | 1,3-beta-glucan synthase component. This family consists of various 1,3-beta-glucan synthase components including Gls1, Gls2 and Gls3 from yeast. 1,3-beta-glucan synthase EC:2.4.1.34 also known as callose synthase catalyzes the formation of a beta-1,3-glucan polymer that is a major component of the fungal cell wall. The reaction catalyzed is:- UDP-glucose + {(1,3)-beta-D-glucosyl}(N) <=> UDP + {(1,3)-beta-D-glucosyl}(N+1). |
| 395036 | Sugar_tr | 5.27e-11 | 1431 | 1764 | 68 | 434 | Sugar (and other) transporter. |
| 340874 | MFS_SV2_like | 1.38e-05 | 1431 | 1764 | 58 | 353 | Metazoan Synaptic vesicle glycoprotein 2 (SV2) and related small molecule transporters of the Major Facilitator Superfamily. This family is composed of metazoan synaptic vesicle glycoprotein 2 (SV2) and related small molecule transporters including those that transport inorganic phosphate (Pht), aromatic compounds (PcaK and related proteins), proline/betaine (ProP), alpha-ketoglutarate (KgtP), citrate (CitA), shikimate (ShiA), and cis,cis-muconate (MucK), among others. SV2 is a transporter-like protein that serves as the receptor for botulinum neurotoxin A (BoNT/A), one of seven neurotoxins produced by the bacterium Clostridium botulinum. BoNT/A blocks neurotransmitter release by cleaving synaptosome-associated protein of 25 kD (SNAP-25) within presynaptic nerve terminals. Also included in this family is synaptic vesicle 2 (SV2)-related protein (SVOP) and similar proteins. SVOP is a transporter-like nucleotide binding protein that localizes to neurotransmitter-containing vesicles. The SV2-like family belongs to the Major Facilitator Superfamily (MFS) of membrane transport proteins, which are thought to function through a single substrate binding site, alternating-access mechanism involving a rocker-switch type of movement. |
| 340874 | MFS_SV2_like | 1.52e-05 | 1429 | 1768 | 56 | 349 | Metazoan Synaptic vesicle glycoprotein 2 (SV2) and related small molecule transporters of the Major Facilitator Superfamily. This family is composed of metazoan synaptic vesicle glycoprotein 2 (SV2) and related small molecule transporters including those that transport inorganic phosphate (Pht), aromatic compounds (PcaK and related proteins), proline/betaine (ProP), alpha-ketoglutarate (KgtP), citrate (CitA), shikimate (ShiA), and cis,cis-muconate (MucK), among others. SV2 is a transporter-like protein that serves as the receptor for botulinum neurotoxin A (BoNT/A), one of seven neurotoxins produced by the bacterium Clostridium botulinum. BoNT/A blocks neurotransmitter release by cleaving synaptosome-associated protein of 25 kD (SNAP-25) within presynaptic nerve terminals. Also included in this family is synaptic vesicle 2 (SV2)-related protein (SVOP) and similar proteins. SVOP is a transporter-like nucleotide binding protein that localizes to neurotransmitter-containing vesicles. The SV2-like family belongs to the Major Facilitator Superfamily (MFS) of membrane transport proteins, which are thought to function through a single substrate binding site, alternating-access mechanism involving a rocker-switch type of movement. |
| 340873 | MFS_GLUT_like | 2.85e-05 | 1431 | 1764 | 56 | 360 | Glucose transporters (GLUTs) and other similar sugar transporters of the Major Facilitator Superfamily. This family is composed of glucose transporters (GLUTs) and other sugar transporters including fungal hexose transporters (HXT), bacterial xylose transporter (XylE), plant sugar transport proteins (STP) and polyol transporters (PLT), H(+)-myo-inositol cotransporter (HMIT), and similar proteins. GLUTs, also called Solute carrier family 2, facilitated glucose transporters (SLC2A), are a family of proteins that facilitate the transport of hexoses such as glucose and fructose. There are fourteen GLUTs found in humans; they display different substrate specificities and tissue expression. They have been categorized into three classes based on sequence similarity: Class 1 (GLUTs 1-4, 14); Class 2 (GLUTs 5, 7, 9, and 11); and Class 3 (GLUTs 6, 8, 10, 12, and HMIT). GLUT proteins are comprised of about 500 amino acid residues, possess a single N-linked oligosaccharide, and have 12 transmembrane segments. The GLUT-like family belongs to the Major Facilitator Superfamily (MFS) of membrane transport proteins, which are thought to function through a single substrate binding site, alternating-access mechanism involving a rocker-switch type of movement. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 1843 | 437 | 2290 | |
| 0.0 | 1 | 1854 | 388 | 2327 | |
| 0.0 | 3 | 1842 | 390 | 2270 | |
| 0.0 | 2 | 1830 | 402 | 2276 | |
| 0.0 | 34 | 1843 | 451 | 2221 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.94e-176 | 1 | 1350 | 610 | 1910 | Callose synthase 5 OS=Arabidopsis thaliana OX=3702 GN=CALS5 PE=1 SV=1 |
|
| 8.50e-175 | 34 | 1350 | 665 | 1915 | Callose synthase 7 OS=Arabidopsis thaliana OX=3702 GN=CALS7 PE=3 SV=3 |
|
| 1.46e-172 | 34 | 1350 | 656 | 1903 | Putative callose synthase 6 OS=Arabidopsis thaliana OX=3702 GN=CALS6 PE=3 SV=2 |
|
| 4.47e-172 | 32 | 1350 | 642 | 1875 | Callose synthase 9 OS=Arabidopsis thaliana OX=3702 GN=CALS9 PE=2 SV=2 |
|
| 2.66e-170 | 34 | 1350 | 652 | 1936 | Callose synthase 3 OS=Arabidopsis thaliana OX=3702 GN=CALS3 PE=3 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.998998 | 0.001020 |
TMHMM Annotations download full data without filtering help
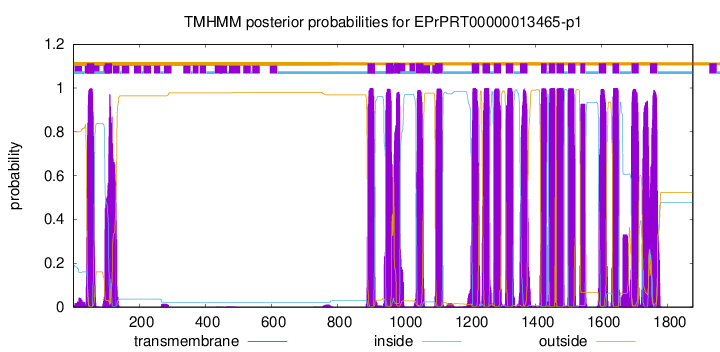
| Start | End |
|---|---|
| 41 | 63 |
| 96 | 118 |
| 891 | 913 |
| 946 | 968 |
| 972 | 989 |
| 1038 | 1060 |
| 1096 | 1118 |
| 1207 | 1229 |
| 1239 | 1261 |
| 1273 | 1295 |
| 1310 | 1332 |
| 1353 | 1375 |
| 1416 | 1433 |
| 1440 | 1459 |
| 1463 | 1485 |
| 1498 | 1520 |
| 1535 | 1550 |
| 1591 | 1613 |
| 1633 | 1652 |
| 1690 | 1712 |
| 1746 | 1768 |
