You are browsing environment: FUNGIDB
CAZyme Information: EPrPRT00000013138-p1
You are here: Home > Sequence: EPrPRT00000013138-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
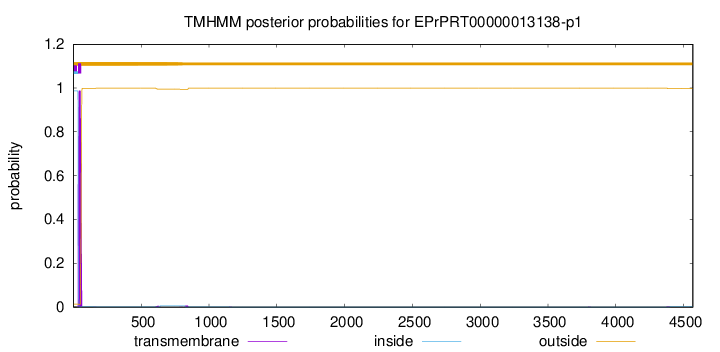
TMHMM annotations
Basic Information help
| Species | Pythium arrhenomanes | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Pythiaceae; Pythium; Pythium arrhenomanes | |||||||||||
| CAZyme ID | EPrPRT00000013138-p1 | |||||||||||
| CAZy Family | AA17 | |||||||||||
| CAZyme Description | Phosphatidylinositol kinase (PIK-L3). | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 67841; End:84340 Strand: - | |||||||||||
Full Sequence Download help
| MDVSNGPAND AESALPLVKE PVAVDNADEQ RRRKLVVIAL SLLALVGLLA LSEISTGGYA | 60 |
| FGSSRDTRSF RDDKKIVVLL SSGRDSQSCA RALVAAKDLA FRSSRVHFRV YEEIAVHHDD | 120 |
| SCLQVFCELS PRDCKALLRT KRLQVTRRDV SGSLGASVGL HVLEGMVDRK SFENDFYLAV | 180 |
| DANIEFTKHW DLELLKQWYS IGNDRAILSV SPPAIEIKGM NHGMLFLQCS ARIHSKDPDP | 240 |
| VVEFNPPEPK PLVKGDSLGA ALLATVPVLQ AQYSERFHFG LVSSLLSVRS DPHLAHLVVG | 300 |
| HEYMRATRFW TKGFDFYAPA RDILYYRYEL PLAPHEKGET DAAIGMSSRR IRRLLKLPLS | 360 |
| SPLFEFELLP DVHILNRLPS NDALRPYVPQ LLHLLLEVLY RDNEDNALLA LKTVFDLHRN | 420 |
| YRPGLRSEVQ AFLDLVQQMY KNVHVTVRKQ FAEAVVTPPS SQAAASPTVS TVATTPTTTA | 480 |
| AASAASTVET SGTTPPSVDG AVSTLSTAST TETTSDPAAT TTTAEPVMPV KTESASAPLS | 540 |
| IAAVSSESPA AAVDTKADGS APPQPPSAAP GVAAATPSTP VSSIASSTVA VDEPLWSSME | 600 |
| SFKAISELPL IIMLLFQCYP TFIESHVPVL VPLMMNLLSL RAPDSAPRQH PQRYVDFLDC | 660 |
| QVKTLSFVTY LLRGCATLMR PFQDAICEST VKLLTVCPKD AFVLRKDIFV AARHIISTEF | 720 |
| RRGFYAQLEL LMDDDVLIGK GRCSFHQIRP LAYSTLADMI HHVRDMLTLS QVSHIVDFYG | 780 |
| KRIHDATLPI SIQTTSIRLL LNLVDISAKN DDADGWKGRT ILSRILLIIA SKFGSTLASL | 840 |
| PVAIGLSLRS AADRGGAGAA PTADLLDAGA MDKIRQSTLL PKAVVEKAPH EKKLDALLAP | 900 |
| YLNAVPRSAS GSVDSFLADE EPCIRDVKSL LRTMILGVRA VIWCTANYRN PHAKDLTSMD | 960 |
| ANASHVSGSD ISAALQGTAT ESSGGIASRP SSAGSTNADA TSAAHAYPLT DDERKLISRV | 1020 |
| LQNGLRCSIL YTLSENTLAE EKQMLDHFAG AFTVLDAPDF RDLFLANMEL LYECILQDHA | 1080 |
| ILTVPQHFLA NSNVSCWFAE ILLKFLITQM KDLSVAAEGD VPDMKREHKV LEMDSLRFER | 1140 |
| MRVVKQEHRA SIVLRLFKIV FGSVTLFKSN ESALFPHLRT IIESCLKEAT FTKHPDNYLL | 1200 |
| LLRALFRSIS GGKYENFYKE VFPLLPGILS ALMRLQQHIQ KPSMHEVLLE LCLTIPARLS | 1260 |
| SLLQYLPSLM KSVIGGRNRQ YQMDPLELAF ESEPQRRALV MEFAWRGLDV NKKPVALGLH | 1320 |
| MDVLLRRAAA ILHRYHRKAP HEWEHVVRKD ARLSDSDEYD DHANTAGATA GGAVVEASLA | 1380 |
| ASAGGGSASS SLLLEKDDKT LELERQAAAN VKQTILQYKR YAFDVVASAT TIAMQLSEAE | 1440 |
| EPDEDEDEEN DEEEEEDDAM NGAETSMRPS VVWESELPTR QLLLKTLFET MDDIDFGEAA | 1500 |
| TRLLMRTATQ MTKKVSDVCD RHEPNFAALS TLQPSSLLPG NRGLSPGRHL ATAKRIEVLR | 1560 |
| QASYGTPFVV VPHSRAAAFF QAFASAILGG LSSPNSQVVD GAKKVLERVV ETALHHYEAD | 1620 |
| IDAASAAYKG GALFLTLTDV FAHACFDKSS WRVKLGGVTG LRLLVDLLAA PFCHENELVI | 1680 |
| VKALFFVLSD HPSEVSATVS VETGEALLAV VDKARDVRGL DIEAFSSGAI KRGDFMSCTA | 1740 |
| FQETEIFQMF VVEFLSPKAA ARRCARQCIA RVAAFTGTTE SAMLFPYQQL ISKQITGCPL | 1800 |
| RSLSSATRIG YIDAMAHALS IQPTIFPLTK ELMTFLQDVW RLISDDATPS ATEAATTAAP | 1860 |
| GIAGGATSTS SNVPVSAQEY PFGLSQASEL RIAAVKLFRA AFLAAPDDMN QHHEARNRFV | 1920 |
| GVFFRYLTRQ PLELVTCAQQ ALSDVIQLNK QHKEITLPKE LLQQCLRPVL LNLADYRKLN | 1980 |
| LPLLDGLSRL LTLLSSCFNV TLGEKLLEHL KQWRDPDRIM KAGIWKRGDE PAVAAAIVDL | 2040 |
| FHLLPPNESF LESLVDCVVE LEAVLPQYGS YGKMSSPYRL PLTRFLNRYA STSIAFFLKR | 2100 |
| DKLIEPKYSS LFQELLKLPE ALPLRNILLS ETGAESLMAA TFVAASKTED IKMQAQIQLN | 2160 |
| AHKAAAQAVA NAQAQGLSPS AAEARGMQAR AAYVAKATAQ VNAQQALKVQ SQVQIQANAQ | 2220 |
| KLHAQTLAAA QAQGLSLVQA QAKAQFASKD YIAKAQSQVA ASTALPSPIS SSAVTPSLAN | 2280 |
| TASPHHVTQL QAQQLHAQIQ VNAQKVHAQA LAAAHAQGLK PIQAQEKAKQ AQATYVHRAK | 2340 |
| AQAQAKIARA AQTSSLSSAL PTTPSALSTV APFTSKAQQE ALELLYQGLR LIRFHYESKL | 2400 |
| LVKILVTYCR AKPDDVQVLL DLLSAFVHRS SSFDNSVLQL FYQQHVAHAY SPYNKRQIVH | 2460 |
| AMQVLIMPVL TTSFDDPTVN NTDMLDADTV QWMLREILAS KDTPTGSPPS DAVLTLRIEL | 2520 |
| LKLGTLLIQH MSKYVTEHRK EVIKFAWNHL KAHDLTSKLW AYVNVCRFIS VYDTPPKIVL | 2580 |
| QVYVALLRTH EMDARFLVRK AFDILLPALP SRLPPNEFIK AIKWTKKIAY EEGHQLGQLV | 2640 |
| HIWFLIVRHP ALFYPFRGQF VPLMVNSLNR LAIPPSSTPD NRRLAVNIVD LIIAWEHTRR | 2700 |
| ERVASRPVTP AAAGDGAALK RRALQAAAGE EETKSESESP SQKRRKVMSG TSEDASAVAV | 2760 |
| AANGSVAASS PTAKTAAAAY SEDDFELNAP MIDLVLNFAF RFALASADKQ ETSRLSKTCG | 2820 |
| ELFDKALRLW PSASVRFSYF DKLIAVTAEA ILRQQPLPTP TPPTQSMPTF FAVPKGAPLS | 2880 |
| SLAILDAVLG ILNALITPEV VSHSARPVPY VVQYAPRVMK LLEPCFDRAN GDIQTGLTTF | 2940 |
| LRRMVELYPP GRAVSQELSA CKFYPWLQEI VADRLQSAVA LQQEFLTSLP SHAASSAVAS | 3000 |
| GSSGGGATQK TKNKQGGGST AAAATASGVS SPASVKSEPK AGQASPGTAA AAGSPAKRQK | 3060 |
| TAGSDPAVAL KVKAALAAAQ SARSSGIPPS KKEPGDKHKK KSSHGEKESA GFAAGTKAGS | 3120 |
| TASSSGVKKK GVAAQHATSG LLSHKMAVEL LIVTLRLLSK CSLPSADHRR VYTNLLIHCL | 3180 |
| ESSTNDKMTL LTKMSSFERL HEVAARPLYD EYYRLVLKLC DPSSDVKHSY FHITPSLPQS | 3240 |
| HSLTTHFVAG LLAPDAEIRE QFLKYLYEAA ASGSPDAAED SLPTGRLLLV LRQDWQACGS | 3300 |
| RYWIAVAVET MLSAATNSTP TGSPRANSKR RLSGQLSPTS PSPSSQLDLL RALRYLAHVD | 3360 |
| VEIAEELWIQ LFRETWQQLS TPQQAHVSSQ LVKTFSSKFN KRDFSAMRLC EFQVENASLR | 3420 |
| VESRLRWIEA LSSIYKTLSD DDLRVGLSLE HIAQPETRAA LVLESLGCVH EAQEEYFKAL | 3480 |
| SKAQSGRVSM DDVNLFELRL WEERWVACAK QLCQWQLMHD FAKATQNQDL LLDCVWKRGD | 3540 |
| WHGAKQLLNF VEVKESIQMM YDIKHASQLH TLPNLKPSIN TWRERLPNKW EPILMWDDIL | 3600 |
| TWRSHMFQVV KNTFSWSDAQ MLACMHDSPW SVIRLAHTAR KQHLPDVCLG ALSKLYTIPA | 3660 |
| MDVQDAFSKL REQVSICYES ASEYTGGLAI LNNTNLDYFS LRQKAEMFRL KALFLEAMGN | 3720 |
| STDANQTFSH CLQICDSYGK GWLSFASQTI ACYLQAIHHR CNAARLMIAR VLWLLSMDDP | 3780 |
| HRGVLIQAFE AHGKQLPIWI WIVWIPQLLM ALGRPEAPQI RGLLRGLSAK FPQALYYTMR | 3840 |
| AFFLENRDSS MAAQQQQMSG QGQVSGSAPT TPQSSGSTGA ATTVTASTGG PGTPTFSGTP | 3900 |
| VYYRTKTGQV VAVPSTMPLN HVQQIPGLVA PARPTPSAFG VALSQVMTVD AWRERLLLGS | 3960 |
| GGADGASSQA TSSSSSSSSA ASSSMPTKPE LGPVQYTEDL LNFLRRTHDS LTFEMECVLE | 4020 |
| EMITKFRPEP EEELLTAVHS LLLKCYQLPR LSQREPVPRM LRAALEKVCR KLFAVTAAST | 4080 |
| TTGSAVGAPP PPQQHKNEKY VEFVAEFKAA FEADFAPDAS LLSAPIQEER VTLFEMMDRL | 4140 |
| KKWRNLLQAR VQHQSCGYST GRASTLRLEH CSRFLMELSS SNMEVPGQYI ADREPIKDLH | 4200 |
| ARIQHFDNDV DVLLRSGYTQ RRLTMGGSDG ATYSFLVQYA MTHTTRTDER MAQMYLLLNR | 4260 |
| LLARHKETRK RNAVFHVPRV VPLTPRSCRA SDTDPDLPME LYRQRISEAY AAMAMAAQHD | 4320 |
| ASETISPQQE DEQLALAKAR AFDEVCASHV PETLLAKYIF ERSAHADAYF EFRSEFTKHL | 4380 |
| ALSSLLAYVL FVGDRAPHRI LFSRRTARVV SAELRPGYAS SGLLEASTSM PFRLTRNLHN | 4440 |
| FVTRPGVHGP FSALMGEEEL LGNQLGLFFR DDLLSWHASK TRLAATASSA GMADSASQQQ | 4500 |
| QQQRQRRLES QVQQRVDANV SLVLERIRGV SLKKETDSQR GKSVRELLEI ATSPERQREM | 4560 |
| YPTWCPWL | 4568 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT60 | 76 | 357 | 1.2e-31 | 0.8727272727272727 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 270707 | PIKK_TRRAP | 7.83e-85 | 4203 | 4478 | 1 | 252 | Pseudokinase domain of TRansformation/tRanscription domain-Associated Protein. TRRAP belongs to the the phosphoinositide 3-kinase-related protein kinase (PIKK) subfamily. It contains a FATC (FRAP, ATM and TRRAP, C-terminal) domain and has a large molecular weight. Unlike most PIKK proteins, however, it contains an inactive PI3K-like pseudokinase domain, which lacks the conserved residues necessary for ATP binding and catalytic activity. TRRAP also contains many motifs that may be critical for protein-protein interactions. TRRAP is a common component of many histone acetyltransferase (HAT) complexes, and is responsible for the recruitment of these complexes to chromatin during transcription, replication, and DNA repair. TRRAP also exists in non-HAT complexes such as the p400 and MRN complexes, which are implicated in ATP-dependent remodeling and DNA repair, respectively. The TRRAP pseudokinase domain subfamily is part of a larger superfamily that includes the catalytic domains of other kinases such as the typical serine/threonine/tyrosine protein kinases (PKs), aminoglycoside phosphotransferase, choline kinase, and RIO kinases. |
| 227365 | TEL1 | 1.57e-38 | 3379 | 4568 | 1073 | 2105 | Phosphatidylinositol kinase or protein kinase, PI-3 family [Signal transduction mechanisms]. |
| 396714 | FAT | 1.47e-27 | 3556 | 3773 | 80 | 342 | FAT domain. The FAT domain is named after FRAP, ATM and TRRAP. |
| 270708 | PIKKc | 3.83e-25 | 4203 | 4455 | 1 | 198 | Catalytic domain of Phosphoinositide 3-kinase-related protein kinases. PIKK subfamily members include ATM (Ataxia telangiectasia mutated), ATR (Ataxia telangiectasia and Rad3-related), TOR (Target of rapamycin), SMG-1 (Suppressor of morphogenetic effect on genitalia-1), and DNA-PK (DNA-dependent protein kinase). PIKKs have intrinsic serine/threonine kinase activity and are distinguished from other PKs by their unique catalytic domain, similar to that of lipid PI3K, and their large molecular weight (240-470 kDa). They show strong preference for phosphorylating serine/threonine residues followed by a glutamine and are also referred to as (S/T)-Q-directed kinases. They all contain a FATC (FRAP, ATM and TRRAP, C-terminal) domain. PIKKs have diverse functions including cell-cycle checkpoints, genome surveillance, mRNA surveillance, and translation control. The PIKK catalytic domain subfamily is part of a larger superfamily that includes the catalytic domains of other kinases such as the typical serine/threonine/tyrosine protein kinases (PKs), aminoglycoside phosphotransferase, choline kinase, and RIO kinases. |
| 270713 | PIKKc_TOR | 9.43e-22 | 4203 | 4476 | 1 | 277 | Catalytic domain of Target of Rapamycin. TOR contains a rapamycin binding domain, a catalytic domain, and a FATC (FRAP, ATM and TRRAP, C-terminal) domain at the C-terminus. It is also called FRAP (FK506 binding protein 12-rapamycin associated protein). TOR is a central component of the eukaryotic growth regulatory network. It controls the expression of many genes transcribed by all three RNA polymerases. It associates with other proteins to form two distinct complexes, TORC1 and TORC2. TORC1 is involved in diverse growth-related functions including protein synthesis, nutrient use and transport, autophagy and stress responses. TORC2 is involved in organizing cytoskeletal structures. TOR is a member of the phosphoinositide 3-kinase-related protein kinase (PIKK) subfamily. PIKKs have intrinsic serine/threonine kinase activity and are distinguished from other PKs by their unique catalytic domain, similar to that of lipid PI3K, and their large molecular weight (240-470 kDa). The TOR catalytic domain subfamily is part of a larger superfamily that includes the catalytic domains of other kinases such as the typical serine/threonine/tyrosine protein kinases (PKs), aminoglycoside phosphotransferase, choline kinase, and RIO kinases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| UIZ27043.1|GT60 | 2.35e-88 | 67 | 360 | 108 | 404 |
| CCA22278.1|GT60 | 3.54e-53 | 73 | 354 | 34 | 306 |
| UIZ22347.1|GT60 | 9.43e-30 | 67 | 365 | 51 | 336 |
| CCA18910.1|GT60 | 4.57e-27 | 74 | 365 | 85 | 364 |
| UIZ25379.1|GT60 | 4.25e-18 | 72 | 367 | 68 | 362 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5OEJ_B | 6.00e-205 | 372 | 4568 | 79 | 3825 | Chain B, Tra1 subunit within the chromatin modifying complex SAGA [Komagataella pastoris],6TB4_L Chain L, Transcription-associated protein [Komagataella phaffii GS115],6TBM_L Chain L, Transcription-associated protein [Komagataella phaffii GS115] |
| 6IG9_T | 1.32e-170 | 374 | 4567 | 82 | 3743 | Tra1 subunit from Saccharomyces cerevisiae SAGA complex [Saccharomyces cerevisiae],6T9I_T cryo-EM structure of transcription coactivator SAGA [Saccharomyces cerevisiae S288C],6T9J_T SAGA Tra1 module [Saccharomyces cerevisiae S288C] |
| 5OJS_T | 1.34e-170 | 374 | 4567 | 105 | 3766 | Cryo-EM structure of the SAGA and NuA4 coactivator subunit Tra1 [Saccharomyces cerevisiae S288C] |
| 7KTS_A | 4.85e-106 | 372 | 2128 | 94 | 1585 | Chain A, Transformation/transcription domain-associated protein,Transformation/transcription domain-associated protein,Transformation/transcription domain-associated protein,Transformation/transcription domain-associated protein,Transformation/transcription domain-associated protein [Homo sapiens] |
| 5Y81_B | 1.37e-96 | 3429 | 4567 | 31 | 1114 | NuA4 TEEAA sub-complex [Saccharomyces cerevisiae S288C] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| sp|Q9Y4A5|TRRAP_HUMAN | 7.81e-198 | 372 | 4568 | 94 | 3859 | Transformation/transcription domain-associated protein OS=Homo sapiens OX=9606 GN=TRRAP PE=1 SV=3 |
| sp|A0A0R4ITC5|TRRAP_DANRE | 1.51e-196 | 372 | 4568 | 94 | 3841 | Transformation/transcription domain-associated protein OS=Danio rerio OX=7955 GN=trrap PE=3 SV=1 |
| sp|P38811|TRA1_YEAST | 6.77e-170 | 374 | 4567 | 82 | 3743 | Transcription-associated protein 1 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=TRA1 PE=1 SV=1 |
| sp|Q8I8U7|TRA1_DROME | 1.37e-168 | 372 | 4568 | 85 | 3790 | Transcription-associated protein 1 OS=Drosophila melanogaster OX=7227 GN=Nipped-A PE=1 SV=4 |
| sp|Q9HFE8|TRA1_SCHPO | 2.66e-158 | 565 | 4568 | 198 | 3699 | Transcription-associated protein 1 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=tra1 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
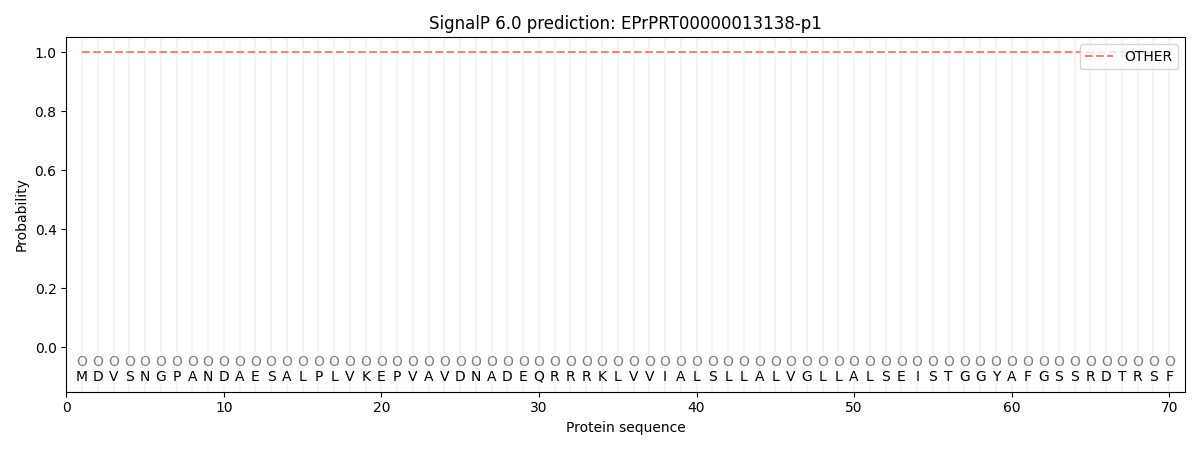
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000020 | 0.000020 |