You are browsing environment: FUNGIDB
CAZyme Information: EPrPIT00000024743-p1
You are here: Home > Sequence: EPrPIT00000024743-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Globisporangium irregulare | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Pythiaceae; Globisporangium; Globisporangium irregulare | |||||||||||
| CAZyme ID | EPrPIT00000024743-p1 | |||||||||||
| CAZy Family | GT31|GT10 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA17 | 26 | 257 | 8.5e-73 | 0.9183673469387755 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.16e-79 | 31 | 203 | 17 | 190 | |
| 5.67e-79 | 15 | 203 | 1 | 190 | |
| 3.10e-77 | 20 | 203 | 6 | 190 | |
| 4.39e-77 | 9 | 203 | 58 | 250 | |
| 1.43e-76 | 30 | 203 | 14 | 188 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.21e-25 | 35 | 201 | 1 | 172 | Chain A, Lytic Polysaccharide Monooxygenase [Phytophthora infestans T30-4],6Z5Y_B Chain B, Lytic Polysaccharide Monooxygenase [Phytophthora infestans T30-4] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
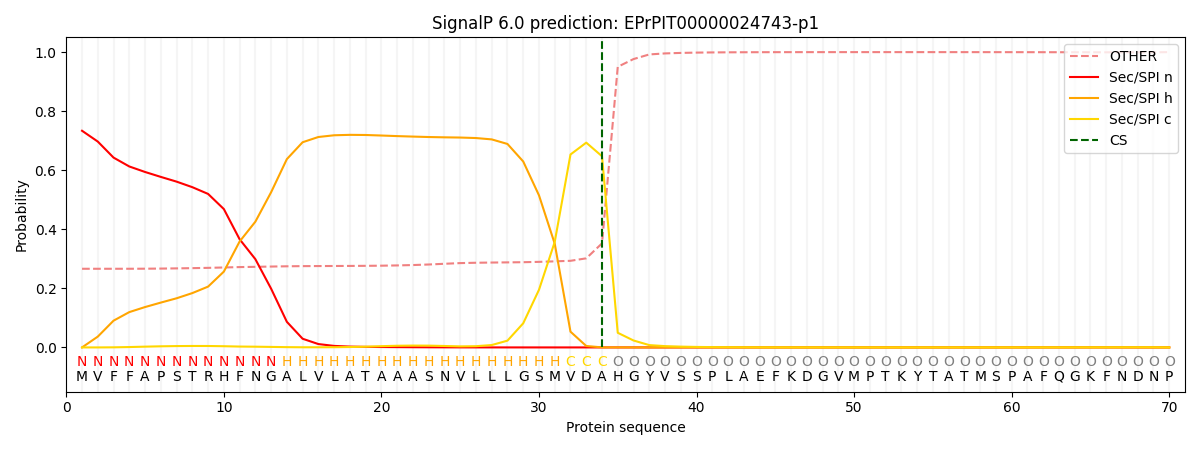
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.274670 | 0.725302 | CS pos: 34-35. Pr: 0.6474 |
