You are browsing environment: FUNGIDB
CAZyme Information: EPrPIT00000023689-p1
You are here: Home > Sequence: EPrPIT00000023689-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Globisporangium irregulare | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Pythiaceae; Globisporangium; Globisporangium irregulare | |||||||||||
| CAZyme ID | EPrPIT00000023689-p1 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | Thioredoxin reductase 1 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA17 | 2 | 202 | 1.5e-44 | 0.8571428571428571 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 282904 | Herpes_BLLF1 | 1.66e-05 | 158 | 204 | 524 | 570 | Herpes virus major outer envelope glycoprotein (BLLF1). This family consists of the BLLF1 viral late glycoprotein, also termed gp350/220. It is the most abundantly expressed glycoprotein in the viral envelope of the Herpesviruses and is the major antigen responsible for stimulating the production of neutralising antibodies in vivo. |
| 237019 | PRK11907 | 0.001 | 157 | 204 | 45 | 92 | bifunctional 2',3'-cyclic-nucleotide 2'-phosphodiesterase/3'-nucleotidase. |
| 236792 | PRK10905 | 0.002 | 157 | 206 | 186 | 232 | cell division protein DamX; Validated |
| 183854 | PRK13042 | 0.003 | 157 | 207 | 44 | 94 | superantigen-like protein SSL4; Reviewed. |
| 237019 | PRK11907 | 0.007 | 157 | 204 | 32 | 84 | bifunctional 2',3'-cyclic-nucleotide 2'-phosphodiesterase/3'-nucleotidase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.24e-49 | 1 | 152 | 27 | 197 | |
| 3.33e-49 | 1 | 152 | 27 | 197 | |
| 3.33e-49 | 1 | 152 | 27 | 197 | |
| 3.34e-47 | 1 | 152 | 27 | 197 | |
| 4.12e-47 | 1 | 157 | 43 | 218 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as OTHER
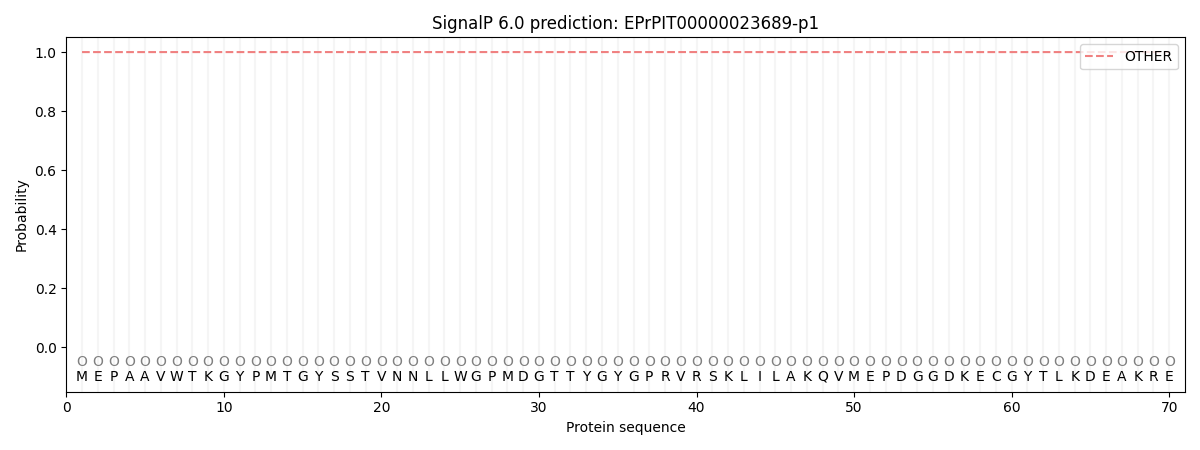
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000072 | 0.000000 |
