You are browsing environment: FUNGIDB
CAZyme Information: EPrPIT00000022548-p1
You are here: Home > Sequence: EPrPIT00000022548-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Globisporangium irregulare | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Pythiaceae; Globisporangium; Globisporangium irregulare | |||||||||||
| CAZyme ID | EPrPIT00000022548-p1 | |||||||||||
| CAZy Family | GH7 | |||||||||||
| CAZyme Description | Chitin-binding protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA16 | 50 | 163 | 6.9e-18 | 0.6766467065868264 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 410624 | LPMO_AA10 | 0.001 | 64 | 132 | 81 | 153 | lytic polysaccharide monooxygenase (LPMO) auxiliary activity family 10 (AA10). AA10 proteins are copper-dependent lytic polysaccharide monooxygenases (LPMOs), which may act on chitin or cellulose. The family used to be called CBM33. Activities in this family include lytic cellulose monooxygenase (C1-hydroxylating) (EC 1.14.99.54), lytic cellulose monooxygenase (C4-dehydrogenating) (EC 1.14.99.56), lytic chitin monooxygenase (EC 1.14.99.53), and lytic xylan monooxygenase/xylan oxidase (glycosidic bond-cleaving) (EC 1.14.99.-). Also included are viral chitin-binding glycoproteins such as fusolin and spheroidin-like proteins. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.68e-13 | 54 | 172 | 73 | 192 | |
| 5.03e-13 | 54 | 172 | 73 | 192 | |
| 5.03e-13 | 54 | 172 | 73 | 192 | |
| 5.03e-13 | 54 | 172 | 73 | 192 | |
| 3.51e-12 | 54 | 174 | 73 | 193 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
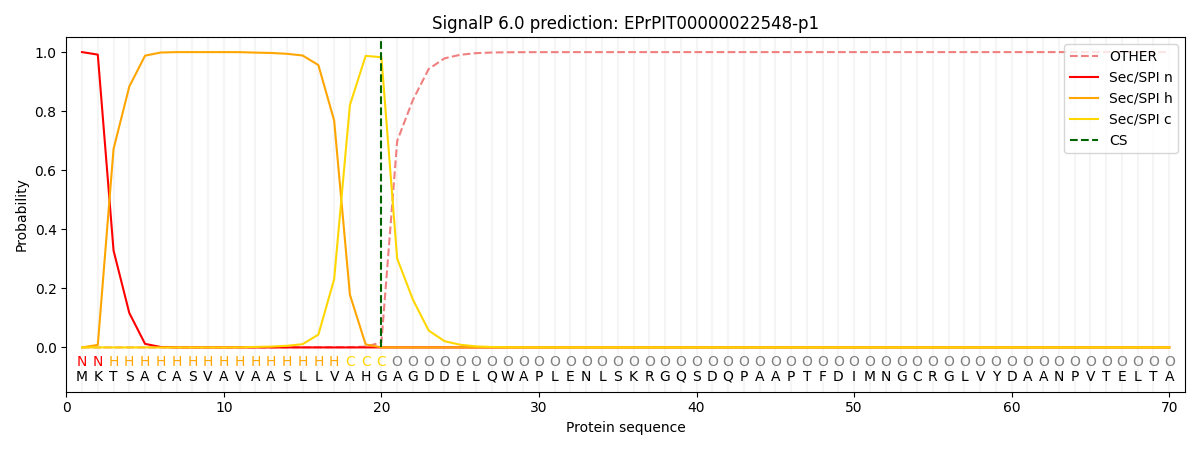
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000296 | 0.999696 | CS pos: 20-21. Pr: 0.9827 |
