You are browsing environment: FUNGIDB
CAZyme Information: EPrPIT00000022339-p1
You are here: Home > Sequence: EPrPIT00000022339-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Globisporangium irregulare | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Pythiaceae; Globisporangium; Globisporangium irregulare | |||||||||||
| CAZyme ID | EPrPIT00000022339-p1 | |||||||||||
| CAZy Family | GH6 | |||||||||||
| CAZyme Description | Dolichyl-phosphate beta-glucosyltransferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT2 | 735 | 910 | 2.6e-25 | 0.9588235294117647 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 240336 | PTZ00260 | 2.26e-112 | 734 | 985 | 72 | 325 | dolichyl-phosphate beta-glucosyltransferase; Provisional |
| 133031 | DPG_synthase | 1.32e-100 | 736 | 955 | 1 | 211 | DPG_synthase is involved in protein N-linked glycosylation. UDP-glucose:dolichyl-phosphate glucosyltransferase (DPG_synthase) is a transmembrane-bound enzyme of the endoplasmic reticulum involved in protein N-linked glycosylation. This enzyme catalyzes the transfer of glucose from UDP-glucose to dolichyl phosphate. |
| 133022 | DPM_DPG-synthase_like | 2.99e-53 | 736 | 934 | 1 | 185 | DPM_DPG-synthase_like is a member of the Glycosyltransferase 2 superfamily. DPM1 is the catalytic subunit of eukaryotic dolichol-phosphate mannose (DPM) synthase. DPM synthase is required for synthesis of the glycosylphosphatidylinositol (GPI) anchor, N-glycan precursor, protein O-mannose, and C-mannose. In higher eukaryotes,the enzyme has three subunits, DPM1, DPM2 and DPM3. DPM is synthesized from dolichol phosphate and GDP-Man on the cytosolic surface of the ER membrane by DPM synthase and then is flipped onto the luminal side and used as a donor substrate. In lower eukaryotes, such as Saccharomyces cerevisiae and Trypanosoma brucei, DPM synthase consists of a single component (Dpm1p and TbDpm1, respectively) that possesses one predicted transmembrane region near the C terminus for anchoring to the ER membrane. In contrast, the Dpm1 homologues of higher eukaryotes, namely fission yeast, fungi, and animals, have no transmembrane region, suggesting the existence of adapter molecules for membrane anchoring. This family also includes bacteria and archaea DPM1_like enzymes. However, the enzyme structure and mechanism of function are not well understood. The UDP-glucose:dolichyl-phosphate glucosyltransferase (DPG_synthase) is a transmembrane-bound enzyme of the endoplasmic reticulum involved in protein N-linked glycosylation. This enzyme catalyzes the transfer of glucose from UDP-glucose to dolichyl phosphate. This protein family belongs to Glycosyltransferase 2 superfamily. |
| 400374 | TBCC | 1.18e-48 | 40 | 158 | 1 | 119 | Tubulin binding cofactor C. Members of this family are involved in the folding pathway of tubulins and form a beta helix structure. |
| 396215 | DHHC | 4.61e-29 | 494 | 615 | 4 | 130 | DHHC palmitoyltransferase. This entry refers to the DHHC domain, found in DHHC proteins which are palmitoyltransferases. Palmitoylation or, more specifically S-acylation, plays important roles in the regulation of protein localization, stability, and activity. It is a post-translational protein modification that involves the attachment of palmitic acid to Cys residues through a thioester linkage. Protein acyltransferases (PATs), also known as palmitoyltransferases, catalyze this reaction by transferring the palmitoyl group from palmitoyl-CoA to the thiol group of Cys residues. They are characterized by the presence of a 50-residue-long domain called the DHHC domain, which in most but not all cases is also cysteine-rich and gets its name from a highly conserved DHHC signature tetrapeptide (Asp-His-His-Cys). The Cys residue within the DHHC domain forms a stable acyl intermediate and transfers the acyl chain to the Cys residues of a target protein. Some proteins containing a DHHC domain include Drosophila DNZ1 protein, Mouse Abl-philin 2 (Aph2) protein, Mammalian ZDHHC9, Yeast ankyrin repeat-containing protein AKR1, Yeast Erf2 protein, and Arabidopsis thaliana tip growth defective 1. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 4.92e-108 | 722 | 986 | 86 | 351 | |
| 3.20e-79 | 722 | 987 | 56 | 321 | |
| 4.31e-79 | 722 | 984 | 55 | 317 | |
| 4.79e-79 | 727 | 984 | 56 | 312 | |
| 4.53e-78 | 728 | 984 | 62 | 321 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.34e-14 | 20 | 171 | 39 | 189 | Crystal Structure of the human Retinitis Pigmentosa protein 2 (RP2) [Homo sapiens] |
|
| 3.40e-14 | 20 | 171 | 41 | 191 | Chain B, Protein XRP2 [Homo sapiens],3BH7_B Crystal structure of the RP2-Arl3 complex bound to GDP-AlF4 [Homo sapiens] |
|
| 1.24e-06 | 727 | 915 | 21 | 192 | Structure of the polyisoprenyl-phosphate glycosyltransferase GtrB (F215A mutant) [Synechocystis sp. PCC 6803 substr. Kazusa],5EKE_B Structure of the polyisoprenyl-phosphate glycosyltransferase GtrB (F215A mutant) [Synechocystis sp. PCC 6803 substr. Kazusa],5EKE_C Structure of the polyisoprenyl-phosphate glycosyltransferase GtrB (F215A mutant) [Synechocystis sp. PCC 6803 substr. Kazusa],5EKE_D Structure of the polyisoprenyl-phosphate glycosyltransferase GtrB (F215A mutant) [Synechocystis sp. PCC 6803 substr. Kazusa] |
|
| 1.24e-06 | 727 | 915 | 21 | 192 | Structure of the polyisoprenyl-phosphate glycosyltransferase GtrB (WT) [Synechocystis sp. PCC 6803 substr. Kazusa],5EKP_B Structure of the polyisoprenyl-phosphate glycosyltransferase GtrB (WT) [Synechocystis sp. PCC 6803 substr. Kazusa],5EKP_C Structure of the polyisoprenyl-phosphate glycosyltransferase GtrB (WT) [Synechocystis sp. PCC 6803 substr. Kazusa],5EKP_D Structure of the polyisoprenyl-phosphate glycosyltransferase GtrB (WT) [Synechocystis sp. PCC 6803 substr. Kazusa] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.25e-78 | 727 | 983 | 61 | 318 | Dolichyl-phosphate beta-glucosyltransferase OS=Drosophila melanogaster OX=7227 GN=wol PE=1 SV=1 |
|
| 4.30e-77 | 720 | 985 | 59 | 327 | Dolichyl-phosphate beta-glucosyltransferase OS=Dictyostelium discoideum OX=44689 GN=alg5 PE=2 SV=1 |
|
| 5.43e-77 | 728 | 985 | 61 | 319 | Dolichyl-phosphate beta-glucosyltransferase OS=Homo sapiens OX=9606 GN=ALG5 PE=1 SV=1 |
|
| 2.54e-73 | 728 | 985 | 61 | 319 | Dolichyl-phosphate beta-glucosyltransferase OS=Mus musculus OX=10090 GN=Alg5 PE=1 SV=1 |
|
| 3.73e-56 | 734 | 982 | 75 | 329 | Dolichyl-phosphate beta-glucosyltransferase OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=ALG5 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000041 | 0.000000 |
TMHMM Annotations download full data without filtering help
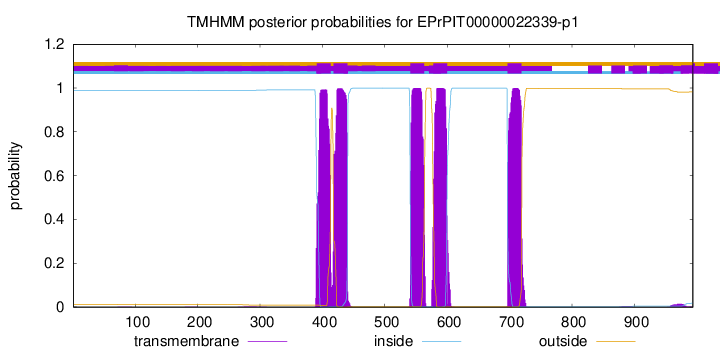
| Start | End |
|---|---|
| 391 | 413 |
| 418 | 440 |
| 541 | 563 |
| 578 | 600 |
| 697 | 719 |
