You are browsing environment: FUNGIDB
CAZyme Information: EPrPIT00000021283-p1
You are here: Home > Sequence: EPrPIT00000021283-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Globisporangium irregulare | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Pythiaceae; Globisporangium; Globisporangium irregulare | |||||||||||
| CAZyme ID | EPrPIT00000021283-p1 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | Polysaccharide lyase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL3 | 86 | 162 | 7.8e-25 | 0.38144329896907214 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397360 | Pectate_lyase | 1.11e-49 | 25 | 163 | 1 | 192 | Pectate lyase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.53e-63 | 4 | 192 | 5 | 261 | |
| 4.85e-58 | 23 | 189 | 31 | 258 | |
| 8.44e-56 | 15 | 191 | 13 | 252 | |
| 4.49e-42 | 28 | 178 | 22 | 236 | |
| 6.37e-33 | 7 | 192 | 12 | 276 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.40e-27 | 29 | 185 | 28 | 240 | Probable pectate lyase G OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=plyG PE=3 SV=1 |
|
| 9.49e-25 | 29 | 185 | 22 | 235 | Probable pectate lyase D OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=plyD PE=3 SV=1 |
|
| 1.91e-24 | 29 | 185 | 23 | 236 | Probable pectate lyase D OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=plyD PE=3 SV=1 |
|
| 1.91e-24 | 29 | 185 | 23 | 236 | Probable pectate lyase D OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=plyD PE=3 SV=1 |
|
| 1.29e-23 | 29 | 185 | 35 | 247 | Probable pectate lyase D OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=plyD PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
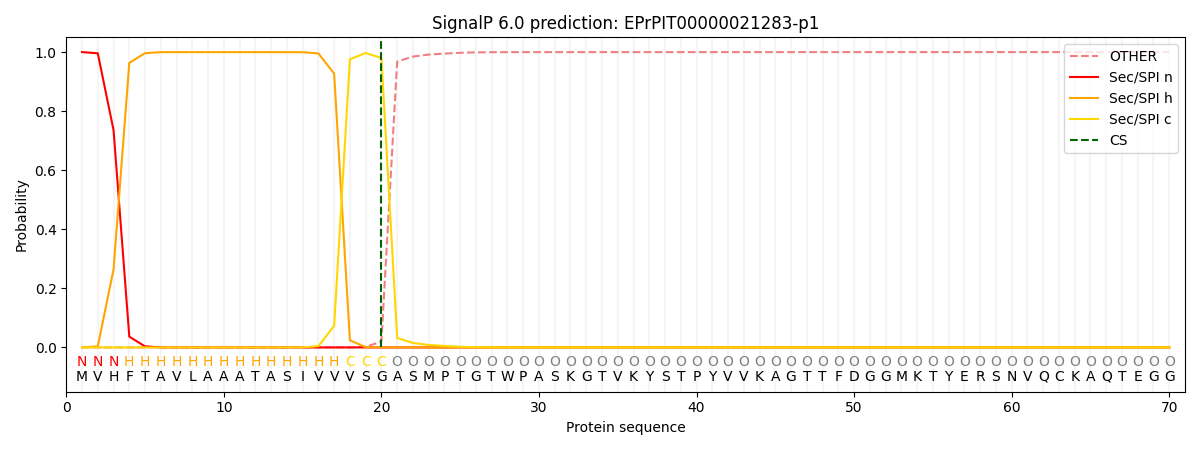
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000189 | 0.999771 | CS pos: 20-21. Pr: 0.9801 |
