You are browsing environment: FUNGIDB
CAZyme Information: EPrPIT00000019551-p1
You are here: Home > Sequence: EPrPIT00000019551-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Globisporangium irregulare | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Pythiaceae; Globisporangium; Globisporangium irregulare | |||||||||||
| CAZyme ID | EPrPIT00000019551-p1 | |||||||||||
| CAZy Family | GH30 | |||||||||||
| CAZyme Description | UDP-N-acetylglucosamine-peptide N-acetylglucosaminyltransferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT41 | 29 | 542 | 2.7e-86 | 0.5801418439716312 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 226428 | Spy | 1.79e-69 | 151 | 553 | 207 | 616 | Predicted O-linked N-acetylglucosamine transferase, SPINDLY family [Posttranslational modification, protein turnover, chaperones]. |
| 404688 | Glyco_transf_41 | 1.22e-65 | 207 | 543 | 81 | 542 | Glycosyl transferase family 41. This family of glycosyltransferases includes O-linked beta-N-acetylglucosamine (O-GlcNAc) transferase, an enzyme which catalyzes the addition of O-GlcNAc to serine and threonine residues. In addition to its function as an O-GlcNAc transferase, human OGT also appears to proteolytically cleave the epigenetic cell-cycle regulator HCF-1. |
| 226975 | COG4627 | 1.21e-30 | 619 | 789 | 7 | 183 | Predicted SAM-depedendent methyltransferase [General function prediction only]. |
| 400514 | Methyltransf_11 | 6.59e-11 | 655 | 700 | 54 | 94 | Methyltransferase domain. Members of this family are SAM dependent methyltransferases. |
| 404528 | Methyltransf_25 | 6.85e-10 | 643 | 696 | 47 | 97 | Methyltransferase domain. This family appears to be a methyltransferase domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.80e-71 | 206 | 789 | 188 | 733 | |
| 2.87e-68 | 206 | 554 | 23 | 380 | |
| 2.00e-67 | 204 | 554 | 14 | 373 | |
| 7.11e-64 | 204 | 554 | 707 | 1066 | |
| 2.00e-63 | 202 | 552 | 558 | 923 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7.00e-48 | 206 | 552 | 234 | 695 | O-GlcNAc transferase from Drososphila melanogaster [Drosophila melanogaster],5A01_B O-GlcNAc transferase from Drososphila melanogaster [Drosophila melanogaster],5A01_C O-GlcNAc transferase from Drososphila melanogaster [Drosophila melanogaster] |
|
| 1.47e-43 | 202 | 552 | 233 | 706 | Crystal structure of the O-GlcNAc transferase Asn648Tyr mutation [Homo sapiens] |
|
| 2.52e-43 | 202 | 552 | 227 | 700 | The human O-GlcNAc transferase in complex with a thiol-linked bisubstrate inhibitor [Homo sapiens] |
|
| 2.54e-43 | 202 | 552 | 228 | 701 | The human O-GlcNAc transferase in complex with a bisubstrate inhibitor [Homo sapiens] |
|
| 2.65e-43 | 202 | 552 | 233 | 706 | Catalytic deficiency of O-GlcNAc transferase leads to X-linked intellectual disability [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.89e-59 | 206 | 552 | 591 | 952 | Probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SEC OS=Arabidopsis thaliana OX=3702 GN=SEC PE=1 SV=1 |
|
| 1.12e-44 | 206 | 570 | 662 | 1147 | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase OS=Caenorhabditis elegans OX=6239 GN=ogt-1 PE=1 SV=2 |
|
| 2.19e-42 | 202 | 552 | 551 | 1024 | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit OS=Mus musculus OX=10090 GN=Ogt PE=1 SV=2 |
|
| 5.15e-42 | 202 | 552 | 551 | 1024 | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit OS=Homo sapiens OX=9606 GN=OGT PE=1 SV=3 |
|
| 5.15e-42 | 202 | 552 | 551 | 1024 | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit OS=Oryctolagus cuniculus OX=9986 GN=OGT PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
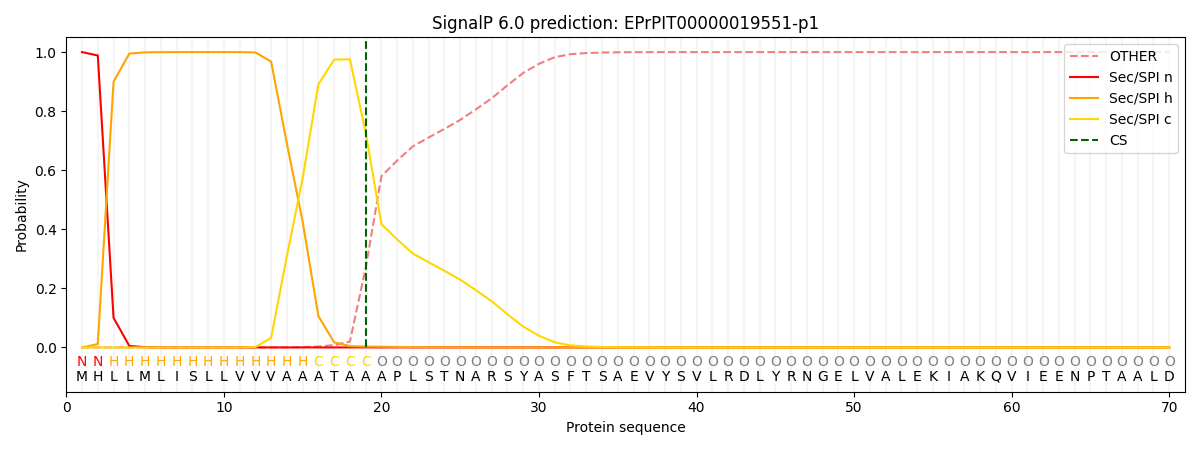
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000249 | 0.999721 | CS pos: 19-20. Pr: 0.7285 |
