You are browsing environment: FUNGIDB
CAZyme Information: EPrPIT00000018377-p1
You are here: Home > Sequence: EPrPIT00000018377-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
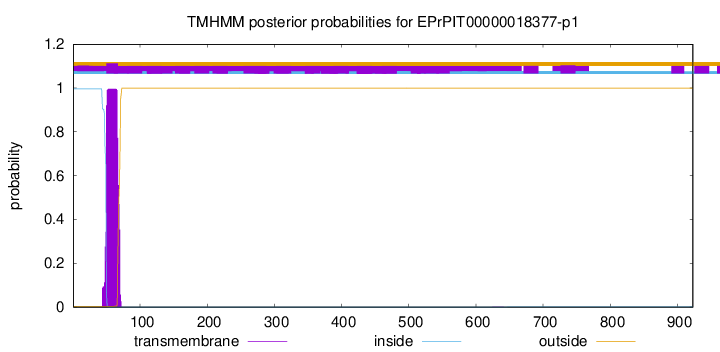
TMHMM annotations
Basic Information help
| Species | Globisporangium irregulare | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Pythiaceae; Globisporangium; Globisporangium irregulare | |||||||||||
| CAZyme ID | EPrPIT00000018377-p1 | |||||||||||
| CAZy Family | GH17 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH35 | 138 | 439 | 4.5e-91 | 0.993485342019544 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 166698 | PLN03059 | 5.01e-122 | 130 | 875 | 29 | 727 | beta-galactosidase; Provisional |
| 396048 | Glyco_hydro_35 | 4.26e-99 | 138 | 438 | 2 | 315 | Glycosyl hydrolases family 35. |
| 224786 | GanA | 1.58e-17 | 131 | 300 | 1 | 171 | Beta-galactosidase GanA [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 6.44e-304 | 20 | 877 | 2 | 796 | |
| 8.15e-239 | 47 | 876 | 40 | 794 | |
| 6.05e-150 | 129 | 875 | 32 | 735 | |
| 5.55e-147 | 129 | 865 | 32 | 721 | |
| 2.14e-142 | 129 | 875 | 32 | 735 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.34e-97 | 129 | 872 | 6 | 703 | Chain A, Beta-galactosidase [Solanum lycopersicum],3W5F_B Chain B, Beta-galactosidase [Solanum lycopersicum],3W5G_A Chain A, Beta-galactosidase [Solanum lycopersicum],3W5G_B Chain B, Beta-galactosidase [Solanum lycopersicum],6IK5_A Chain A, Beta-galactosidase [Solanum lycopersicum],6IK5_B Chain B, Beta-galactosidase [Solanum lycopersicum] |
|
| 2.42e-96 | 129 | 872 | 6 | 703 | Chain A, Beta-galactosidase [Solanum lycopersicum],6IK6_B Chain B, Beta-galactosidase [Solanum lycopersicum],6IK7_A Chain A, Beta-galactosidase [Solanum lycopersicum],6IK7_B Chain B, Beta-galactosidase [Solanum lycopersicum],6IK8_A Chain A, Beta-galactosidase [Solanum lycopersicum],6IK8_B Chain B, Beta-galactosidase [Solanum lycopersicum] |
|
| 2.45e-41 | 131 | 463 | 18 | 357 | Chain A, Beta-galactosidase [Niallia circulans],4MAD_B Chain B, Beta-galactosidase [Niallia circulans] |
|
| 2.73e-38 | 129 | 402 | 9 | 291 | Crystal structure of human beta-galactosidase in complex with galactose [Homo sapiens],3THC_B Crystal structure of human beta-galactosidase in complex with galactose [Homo sapiens],3THC_C Crystal structure of human beta-galactosidase in complex with galactose [Homo sapiens],3THC_D Crystal structure of human beta-galactosidase in complex with galactose [Homo sapiens],3THD_A Crystal structure of human beta-galactosidase in complex with 1-deoxygalactonojirimycin [Homo sapiens],3THD_B Crystal structure of human beta-galactosidase in complex with 1-deoxygalactonojirimycin [Homo sapiens],3THD_C Crystal structure of human beta-galactosidase in complex with 1-deoxygalactonojirimycin [Homo sapiens],3THD_D Crystal structure of human beta-galactosidase in complex with 1-deoxygalactonojirimycin [Homo sapiens] |
|
| 3.36e-38 | 129 | 402 | 33 | 315 | Crystal structure of human beta-galactosidase in complex with NOEV [Homo sapiens],3WEZ_B Crystal structure of human beta-galactosidase in complex with NOEV [Homo sapiens],3WEZ_C Crystal structure of human beta-galactosidase in complex with NOEV [Homo sapiens],3WEZ_D Crystal structure of human beta-galactosidase in complex with NOEV [Homo sapiens],3WF0_A Crystal structure of human beta-galactosidase in complex with 6S-NBI-DGJ [Homo sapiens],3WF0_B Crystal structure of human beta-galactosidase in complex with 6S-NBI-DGJ [Homo sapiens],3WF0_C Crystal structure of human beta-galactosidase in complex with 6S-NBI-DGJ [Homo sapiens],3WF0_D Crystal structure of human beta-galactosidase in complex with 6S-NBI-DGJ [Homo sapiens],3WF1_A Crystal structure of human beta-galactosidase in complex with 6S-NBI-GJ [Homo sapiens],3WF1_B Crystal structure of human beta-galactosidase in complex with 6S-NBI-GJ [Homo sapiens],3WF1_C Crystal structure of human beta-galactosidase in complex with 6S-NBI-GJ [Homo sapiens],3WF1_D Crystal structure of human beta-galactosidase in complex with 6S-NBI-GJ [Homo sapiens],3WF2_A Crystal structure of human beta-galactosidase in complex with NBT-DGJ [Homo sapiens],3WF2_B Crystal structure of human beta-galactosidase in complex with NBT-DGJ [Homo sapiens],3WF2_C Crystal structure of human beta-galactosidase in complex with NBT-DGJ [Homo sapiens],3WF2_D Crystal structure of human beta-galactosidase in complex with NBT-DGJ [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.61e-113 | 126 | 872 | 28 | 709 | Beta-galactosidase 11 OS=Oryza sativa subsp. japonica OX=39947 GN=Os08g0549200 PE=2 SV=1 |
|
| 2.24e-112 | 130 | 876 | 30 | 731 | Beta-galactosidase 5 OS=Arabidopsis thaliana OX=3702 GN=BGAL5 PE=2 SV=1 |
|
| 1.91e-104 | 121 | 873 | 33 | 719 | Beta-galactosidase 14 OS=Arabidopsis thaliana OX=3702 GN=BGAL14 PE=2 SV=2 |
|
| 1.01e-101 | 131 | 870 | 28 | 727 | Beta-galactosidase 7 OS=Arabidopsis thaliana OX=3702 GN=BGAL7 PE=2 SV=2 |
|
| 2.70e-101 | 131 | 876 | 27 | 726 | Beta-galactosidase 5 OS=Oryza sativa subsp. japonica OX=39947 GN=Os03g0165400 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999946 | 0.000064 |