You are browsing environment: FUNGIDB
CAZyme Information: EPrPIT00000015629-p1
You are here: Home > Sequence: EPrPIT00000015629-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Globisporangium irregulare | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Pythiaceae; Globisporangium; Globisporangium irregulare | |||||||||||
| CAZyme ID | EPrPIT00000015629-p1 | |||||||||||
| CAZy Family | AA17 | |||||||||||
| CAZyme Description | Glucan 1,3-beta-glucosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 115 | 397 | 2.3e-52 | 0.9711191335740073 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 225344 | BglC | 1.10e-27 | 65 | 402 | 19 | 369 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| 395098 | Cellulase | 3.66e-16 | 122 | 393 | 24 | 268 | Cellulase (glycosyl hydrolase family 5). |
| 270383 | PBP2_TRAP_Dctp3_4 | 0.009 | 84 | 134 | 217 | 278 | Periplasmic substrate-binding component of TRAP-type C4-dicarboxylate transport system DctP3 and DctP4; the type 2 periplasmic-binding protein fold. This group includes uncharacterized DctP3 and DctP 4 subfamilies of TRAP Transporters specific to C4-dicarboxylates such as succinate, malate and fumarate. TRAP transporters are a large family of solute transporters ubiquitously found in bacteria and archaea. This CD also included some eukaryotic homologs that have not been functionally characterized. TRAP transporters are comprised of a periplasmic substrate-binding protein (SBP; often called the P subunit) and two unequally sized integral membrane components: a large transmembrane subunit involved in the translocation process (the M subunit) and a smaller membrane of unknown function (the Q subunit). The driving force of TRAP transporters is provided by electrochemical ion gradients (either protons or sodium ions) across the cytoplasmic membrane, rather than ATP hydrolysis. This substrate-binding domain belongs to the type 2 periplasmic binding fold protein superfamily (PBP2). The PBP2 proteins are typically comprised of two globular subdomains connected by a flexible hinge and bind their ligand in the cleft between these domains in a manner resembling a Venus flytrap. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.40e-171 | 46 | 419 | 41 | 413 | |
| 1.50e-162 | 36 | 419 | 16 | 406 | |
| 1.02e-116 | 57 | 416 | 5 | 361 | |
| 5.01e-116 | 53 | 416 | 9 | 369 | |
| 2.32e-112 | 46 | 412 | 120 | 491 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.98e-33 | 65 | 409 | 10 | 372 | Exo-b-(1,3)-glucanase From Candida Albicans [Candida albicans] |
|
| 2.97e-33 | 65 | 409 | 15 | 377 | F144Y/F258Y Double Mutant of Exo-beta-1,3-glucanase from Candida albicans at 2 A [Candida albicans] |
|
| 3.77e-33 | 65 | 409 | 10 | 372 | Exo-b-(1,3)-glucanase From Candida Albicans At 1.85 A Resolution [Candida albicans],1EQC_A Exo-b-(1,3)-glucanase From Candida Albicans In Complex With Castanospermine At 1.85 A [Candida albicans] |
|
| 4.16e-33 | 65 | 409 | 16 | 378 | Exo-B-(1,3)-Glucanase from Candida Albicans in complex with unhydrolysed and covalently linked 2,4-dinitrophenyl-2-deoxy-2-fluoro-B-D-glucopyranoside at 1.9 A [Candida albicans] |
|
| 4.16e-33 | 65 | 409 | 16 | 378 | Chain A, Hypothetical protein XOG1 [Candida albicans] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.01e-35 | 63 | 410 | 41 | 405 | Glucan 1,3-beta-glucosidase OS=Schwanniomyces occidentalis OX=27300 PE=3 SV=1 |
|
| 1.14e-34 | 63 | 410 | 48 | 414 | Glucan 1,3-beta-glucosidase OS=Pichia angusta OX=870730 PE=3 SV=1 |
|
| 1.36e-34 | 63 | 411 | 36 | 399 | Glucan 1,3-beta-glucosidase OS=Blumeria graminis OX=34373 PE=3 SV=1 |
|
| 9.55e-34 | 63 | 394 | 41 | 395 | Glucan 1,3-beta-glucosidase 2 OS=Wickerhamomyces anomalus OX=4927 GN=EXG2 PE=3 SV=1 |
|
| 2.01e-32 | 65 | 409 | 54 | 416 | Glucan 1,3-beta-glucosidase OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=XOG1 PE=1 SV=5 |
SignalP and Lipop Annotations help
This protein is predicted as SP
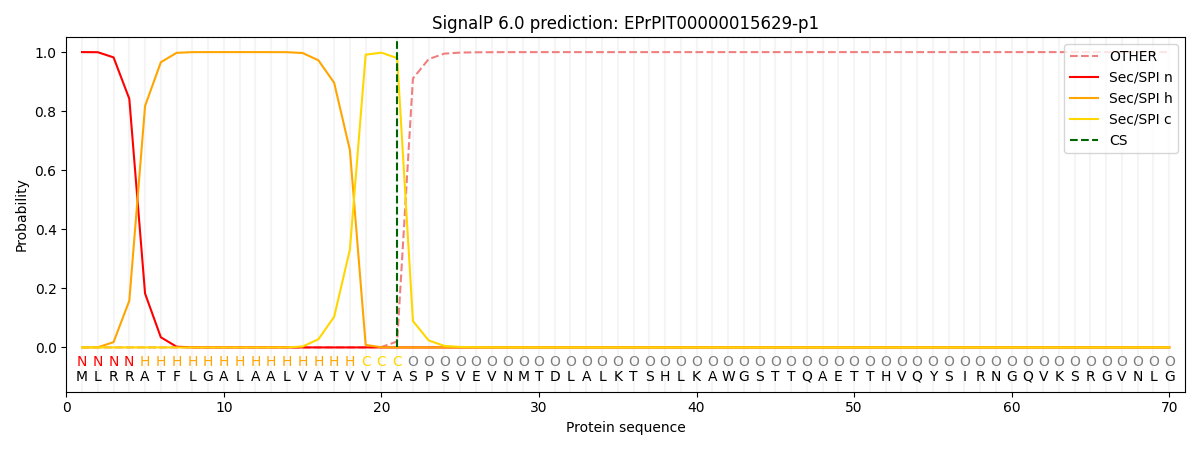
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000296 | 0.999677 | CS pos: 21-22. Pr: 0.9797 |
