You are browsing environment: FUNGIDB
CAZyme Information: EPrPIT00000015134-p1
You are here: Home > Sequence: EPrPIT00000015134-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
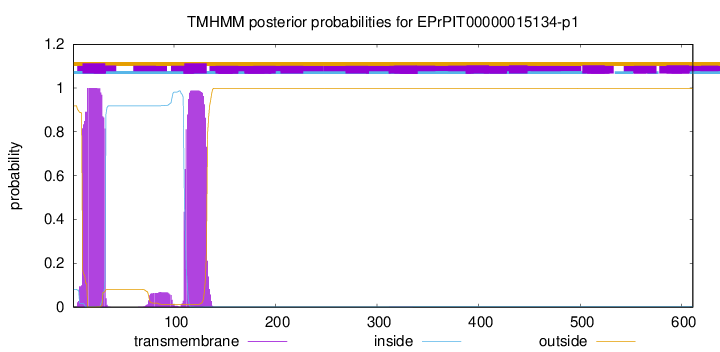
TMHMM annotations
Basic Information help
| Species | Globisporangium irregulare | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Pythiaceae; Globisporangium; Globisporangium irregulare | |||||||||||
| CAZyme ID | EPrPIT00000015134-p1 | |||||||||||
| CAZy Family | AA17 | |||||||||||
| CAZyme Description | Transmembrane protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT110 | 338 | 512 | 9e-17 | 0.8932584269662921 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397245 | Exostosin | 0.003 | 476 | 512 | 217 | 253 | Exostosin family. The EXT family is a family of tumor suppressor genes. Mutations of EXT1 on 8q24.1, EXT2 on 11p11-13, and EXT3 on 19p have been associated with the autosomal dominant disorder known as hereditary multiple exostoses (HME). This is the most common known skeletal dysplasia. The chromosomal locations of other EXT genes suggest association with other forms of neoplasia. EXT1 and EXT2 have both been shown to encode a heparan sulphate polymerase with both D-glucuronyl (GlcA) and N-acetyl-D-glucosaminoglycan (GlcNAC) transferase activities. The nature of the defect in heparan sulphate biosynthesis in HME is unclear. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 4.10e-14 | 311 | 589 | 134 | 387 | |
| 1.49e-13 | 334 | 586 | 165 | 383 | |
| 6.61e-12 | 319 | 597 | 184 | 432 | |
| 9.05e-11 | 320 | 589 | 140 | 376 | |
| 9.37e-11 | 334 | 589 | 160 | 386 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.31e-11 | 319 | 589 | 174 | 411 | Ribitol-5-phosphate xylosyltransferase 1 OS=Danio rerio OX=7955 GN=rxylt1 PE=2 SV=2 |
|
| 2.21e-08 | 477 | 606 | 318 | 438 | Ribitol-5-phosphate xylosyltransferase 1 OS=Homo sapiens OX=9606 GN=RXYLT1 PE=1 SV=1 |
|
| 5.14e-08 | 477 | 589 | 319 | 419 | Ribitol-5-phosphate xylosyltransferase 1 OS=Mus musculus OX=10090 GN=Rxylt1 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000062 | 0.000001 |