You are browsing environment: FUNGIDB
CAZyme Information: EPrPAT00000024497-p1
You are here: Home > Sequence: EPrPAT00000024497-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Pythium aphanidermatum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Pythiaceae; Pythium; Pythium aphanidermatum | |||||||||||
| CAZyme ID | EPrPAT00000024497-p1 | |||||||||||
| CAZy Family | GT71 | |||||||||||
| CAZyme Description | Carbohydrate-binding protein. | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 1.1.99.18:3 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA3 | 19 | 552 | 3.6e-104 | 0.9708029197080292 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 223562 | Uup | 2.75e-55 | 1024 | 1416 | 18 | 309 | ATPase components of ABC transporters with duplicated ATPase domains [General function prediction only]. |
| 215558 | PLN03073 | 2.72e-44 | 996 | 1403 | 164 | 482 | ABC transporter F family; Provisional |
| 225186 | BetA | 2.93e-34 | 26 | 557 | 7 | 535 | Choline dehydrogenase or related flavoprotein [Lipid transport and metabolism, General function prediction only]. |
| 213188 | ABCF_EF-3 | 5.26e-30 | 1249 | 1329 | 68 | 142 | ATP-binding cassette domain of elongation factor 3, subfamily F. Elongation factor 3 (EF-3) is a cytosolic protein required by fungal ribosomes for in vitro protein synthesis and for in vivo growth. EF-3 stimulates the binding of the EF-1: GTP: aa-tRNA ternary complex to the ribosomal A site by facilitated release of the deacylated tRNA from the E site. The reaction requires ATP hydrolysis. EF-3 contains two ATP nucleotide binding sequence (NBS) motifs. NBSI is sufficient for the intrinsic ATPase activity. NBSII is essential for the ribosome-stimulated functions. |
| 223562 | Uup | 5.31e-26 | 1019 | 1350 | 332 | 529 | ATPase components of ABC transporters with duplicated ATPase domains [General function prediction only]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 8.37e-241 | 25 | 886 | 22 | 950 | |
| 2.93e-218 | 25 | 889 | 22 | 885 | |
| 3.91e-202 | 21 | 641 | 23 | 638 | |
| 4.01e-160 | 25 | 550 | 22 | 544 | |
| 2.68e-148 | 1 | 537 | 1 | 526 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.78e-56 | 27 | 551 | 3 | 530 | Chain A, Cellobiose dehydrogenase [Phanerodontia chrysosporium],1NAA_B Chain B, Cellobiose dehydrogenase [Phanerodontia chrysosporium] |
|
| 3.09e-56 | 27 | 551 | 8 | 535 | Chain A, cellobiose dehydrogenase [Phanerodontia chrysosporium],1KDG_B Chain B, cellobiose dehydrogenase [Phanerodontia chrysosporium] |
|
| 1.35e-33 | 26 | 571 | 7 | 558 | Chain A, Cellobiose dehydrogenase [Thermothelomyces myriococcoides],4QI5_A Chain A, Cellobiose dehydrogenase [Thermothelomyces myriococcoides] |
|
| 6.01e-33 | 26 | 571 | 229 | 780 | Chain A, Cellobiose dehydrogenase [Thermothelomyces myriococcoides] |
|
| 5.54e-32 | 26 | 571 | 229 | 776 | Cellobiose dehydrogenase from Neurospora crassa, NcCDH [Neurospora crassa OR74A],4QI7_B Cellobiose dehydrogenase from Neurospora crassa, NcCDH [Neurospora crassa OR74A] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.80e-54 | 27 | 551 | 235 | 762 | Cellobiose dehydrogenase OS=Phanerodontia chrysosporium OX=2822231 GN=CDH-1 PE=1 SV=1 |
|
| 1.36e-30 | 995 | 1384 | 161 | 464 | ABC transporter F family member 3 OS=Arabidopsis thaliana OX=3702 GN=ABCF3 PE=1 SV=1 |
|
| 1.14e-29 | 1002 | 1353 | 196 | 462 | Protein GCN20 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=GCN20 PE=1 SV=1 |
|
| 5.40e-29 | 999 | 1353 | 157 | 413 | ABC transporter F family member 4 OS=Arabidopsis thaliana OX=3702 GN=ABCF4 PE=2 SV=1 |
|
| 3.51e-27 | 1000 | 1353 | 261 | 507 | ATP-binding cassette sub-family F member 1 OS=Sus scrofa OX=9823 GN=ABCF1 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
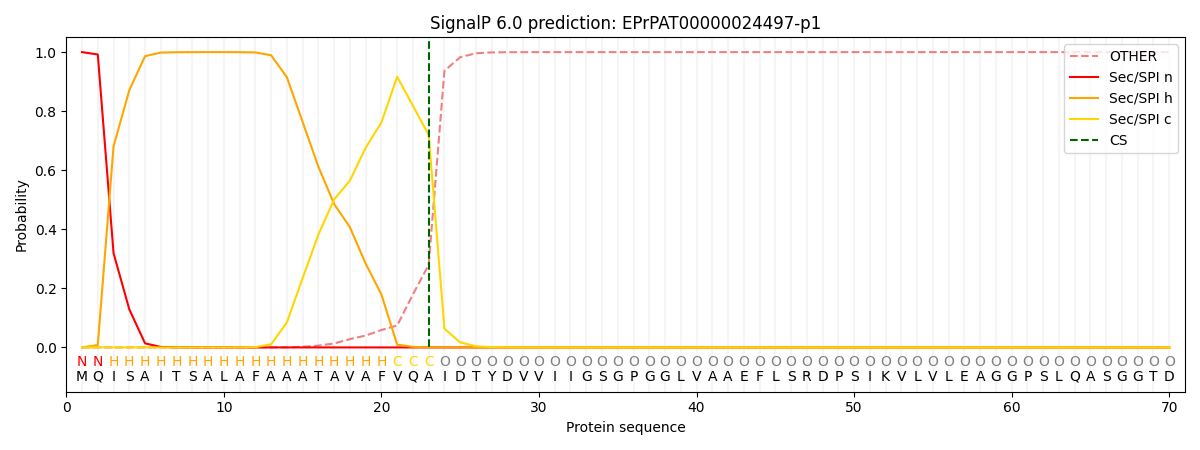
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.001070 | 0.998891 | CS pos: 23-24. Pr: 0.7198 |
