You are browsing environment: FUNGIDB
CAZyme Information: EPrPAT00000022299-p1
You are here: Home > Sequence: EPrPAT00000022299-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Pythium aphanidermatum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Pythiaceae; Pythium; Pythium aphanidermatum | |||||||||||
| CAZyme ID | EPrPAT00000022299-p1 | |||||||||||
| CAZy Family | GT31 | |||||||||||
| CAZyme Description | Aldose 1-epimerase. | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.265:13 | 2.4.1.-:9 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT57 | 334 | 808 | 6.9e-142 | 0.8856548856548857 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397324 | Alg6_Alg8 | 2.59e-154 | 342 | 795 | 10 | 419 | ALG6, ALG8 glycosyltransferase family. N-linked (asparagine-linked) glycosylation of proteins is mediated by a highly conserved pathway in eukaryotes, in which a lipid (dolichol phosphate)-linked oligosaccharide is assembled at the endoplasmic reticulum membrane prior to the transfer of the oligosaccharide moiety to the target asparagine residues. This oligosaccharide is composed of Glc(3)Man(9)GlcNAc(2). The addition of the three glucose residues is the final series of steps in the synthesis of the oligosaccharide precursor. Alg6 transfers the first glucose residue, and Alg8 transfers the second one. In the human alg6 gene, a C->T transition, which causes Ala333 to be replaced with Val, has been identified as the cause of a congenital disorder of glycosylation, designated as type Ic OMIM:603147. |
| 185697 | D-hex-6-P-epi_like | 8.16e-99 | 6 | 291 | 1 | 269 | D-hexose-6-phosphate epimerase-like. D-Hexose-6-phosphate epimerase Ymr099c from Saccharomyces cerevisiae belongs to the large superfamily of aldose-1-epimerases. Its active site is very similar to the catalytic site of galactose mutarotase, the best studied member of the superfamily. It also contains the conserved glutamate and histidine residues that have been shown in galactose mutarotase to be critical for catalysis, the glutamate serving as the active site base to initiate the reaction by removing the proton from the C-1 hydroxyl group of the sugar substrate, and the histidine as the active site acid to protonate the C-5 ring oxygen. In addition Ymr099c contains 2 conserved arginine residues which are involved in phosphate binding, and exhibits hexose-6-phosphate mutarotase activity on glucose-6-P, galactose-6-P and mannose-6-P. |
| 223748 | YeaD | 3.07e-56 | 3 | 293 | 23 | 285 | D-hexose-6-phosphate mutarotase [Carbohydrate transport and metabolism]. |
| 396013 | Aldose_epim | 8.21e-39 | 5 | 291 | 1 | 300 | Aldose 1-epimerase. |
| 185702 | Aldose_epim_Slr1438 | 4.97e-21 | 37 | 269 | 33 | 241 | Aldose 1-epimerase, similar to Synechocystis Slr1438. Proteins similar to Synechocystis Slr1438 are uncharacterized members of aldose-1-epimerase superfamily. Aldose 1-epimerases or mutarotases are key enzymes of carbohydrate metabolism, catalyzing the interconversion of the alpha- and beta-anomers of hexose sugars such as glucose and galactose. This interconversion is an important step that allows anomer specific metabolic conversion of sugars. Studies of the catalytic mechanism of the best known member of the family, galactose mutarotase, have shown a glutamate and a histidine residue to be critical for catalysis; the glutamate serves as the active site base to initiate the reaction by removing the proton from the C-1 hydroxyl group of the sugar substrate, and the histidine as the active site acid to protonate the C-5 ring oxygen. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 4.70e-204 | 318 | 806 | 10 | 503 | |
| 1.38e-126 | 329 | 779 | 15 | 434 | |
| 5.63e-104 | 336 | 787 | 24 | 450 | |
| 5.63e-104 | 336 | 787 | 24 | 450 | |
| 5.63e-104 | 336 | 787 | 24 | 450 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.51e-43 | 5 | 288 | 8 | 284 | Structure-based functional annotation: Yeast ymr099c codes for a D- hexose-6-phosphate mutarotase. [Saccharomyces cerevisiae S288C] |
|
| 4.51e-43 | 5 | 288 | 8 | 284 | Structure-based functional annotation: Yeast ymr099c codes for a D- hexose-6-phosphate mutarotase. Complex with glucose-6-phosphate [Saccharomyces cerevisiae S288C],2CIS_A Structure-based functional annotation: Yeast ymr099c codes for a D- hexose-6-phosphate mutarotase. Complex with tagatose-6-phosphate [Saccharomyces cerevisiae S288C] |
|
| 2.14e-22 | 1 | 290 | 38 | 304 | Chain A, Putative enzyme related to aldose 1-epimerase [Salmonella enterica subsp. enterica serovar Typhimurium],2HTA_B Chain B, Putative enzyme related to aldose 1-epimerase [Salmonella enterica subsp. enterica serovar Typhimurium],2HTB_A Chain A, Putative enzyme related to aldose 1-epimerase [Salmonella enterica subsp. enterica serovar Typhimurium],2HTB_B Chain B, Putative enzyme related to aldose 1-epimerase [Salmonella enterica subsp. enterica serovar Typhimurium],2HTB_C Chain C, Putative enzyme related to aldose 1-epimerase [Salmonella enterica subsp. enterica serovar Typhimurium],2HTB_D Chain D, Putative enzyme related to aldose 1-epimerase [Salmonella enterica subsp. enterica serovar Typhimurium] |
|
| 4.62e-19 | 348 | 757 | 87 | 433 | Cryo-EM structure of yeast ALG6 in complex with 6AG9 Fab and Dol25-P-Glc [Saccharomyces cerevisiae],6SNI_X Cryo-EM structure of nanodisc reconstituted yeast ALG6 in complex with 6AG9 Fab [Saccharomyces cerevisiae] |
|
| 9.89e-15 | 2 | 272 | 19 | 252 | Crystal Structure Analysis of HI1317 [Haemophilus influenzae] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.00e-104 | 336 | 787 | 24 | 450 | Probable dolichyl pyrophosphate Glc1Man9GlcNAc2 alpha-1,3-glucosyltransferase OS=Mus musculus OX=10090 GN=Alg8 PE=2 SV=2 |
|
| 3.86e-98 | 336 | 787 | 24 | 450 | Probable dolichyl pyrophosphate Glc1Man9GlcNAc2 alpha-1,3-glucosyltransferase OS=Bos taurus OX=9913 GN=ALG8 PE=2 SV=1 |
|
| 4.06e-97 | 336 | 787 | 24 | 450 | Probable dolichyl pyrophosphate Glc1Man9GlcNAc2 alpha-1,3-glucosyltransferase OS=Homo sapiens OX=9606 GN=ALG8 PE=1 SV=2 |
|
| 6.41e-96 | 332 | 793 | 14 | 441 | Dolichyl pyrophosphate Glc1Man9GlcNAc2 alpha-1,3-glucosyltransferase OS=Neurospora crassa (strain ATCC 24698 / 74-OR23-1A / CBS 708.71 / DSM 1257 / FGSC 987) OX=367110 GN=alg-8 PE=3 SV=2 |
|
| 9.68e-95 | 330 | 779 | 16 | 421 | Probable dolichyl pyrophosphate Glc1Man9GlcNAc2 alpha-1,3-glucosyltransferase OS=Arabidopsis thaliana OX=3702 GN=At2g44660 PE=2 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000050 | 0.000008 |
TMHMM Annotations download full data without filtering help
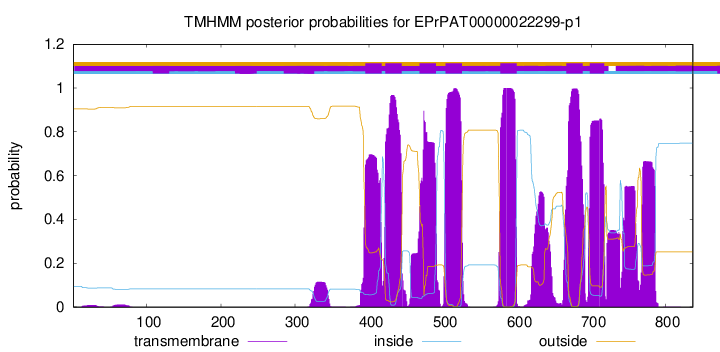
| Start | End |
|---|---|
| 395 | 417 |
| 422 | 444 |
| 468 | 490 |
| 503 | 525 |
| 577 | 599 |
| 666 | 688 |
| 698 | 717 |
