You are browsing environment: FUNGIDB
CAZyme Information: EPrPAT00000021465-p1
You are here: Home > Sequence: EPrPAT00000021465-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Pythium aphanidermatum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Pythiaceae; Pythium; Pythium aphanidermatum | |||||||||||
| CAZyme ID | EPrPAT00000021465-p1 | |||||||||||
| CAZy Family | GT105 | |||||||||||
| CAZyme Description | Callose synthase. | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT48 | 1219 | 1884 | 3.8e-264 | 0.945872801082544 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396784 | Glucan_synthase | 0.0 | 1219 | 1965 | 3 | 816 | 1,3-beta-glucan synthase component. This family consists of various 1,3-beta-glucan synthase components including Gls1, Gls2 and Gls3 from yeast. 1,3-beta-glucan synthase EC:2.4.1.34 also known as callose synthase catalyzes the formation of a beta-1,3-glucan polymer that is a major component of the fungal cell wall. The reaction catalyzed is:- UDP-glucose + {(1,3)-beta-D-glucosyl}(N) <=> UDP + {(1,3)-beta-D-glucosyl}(N+1). |
| 405046 | FKS1_dom1 | 1.18e-25 | 386 | 472 | 6 | 106 | 1,3-beta-glucan synthase subunit FKS1, domain-1. The FKS1_dom1 domain is likely to be the 'Class I' region just N-terminal to the first set of transmembrane helices that is involved in 1,3-beta-glucan synthesis itself. This family is found on proteins with family Glucan_synthase, pfam02364. |
| 404726 | DUF4203 | 4.60e-08 | 47 | 183 | 1 | 146 | Domain of unknown function (DUF4203). This is the N-terminal region of 7tm proteins. The function is not known. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 2160 | 415 | 2605 | |
| 0.0 | 1 | 2108 | 405 | 2531 | |
| 6.51e-255 | 288 | 2045 | 22 | 1716 | |
| 3.06e-253 | 288 | 2045 | 224 | 1918 | |
| 1.52e-248 | 312 | 2045 | 260 | 1930 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 8.26e-242 | 312 | 2044 | 248 | 1921 | Callose synthase 5 OS=Arabidopsis thaliana OX=3702 GN=CALS5 PE=1 SV=1 |
|
| 6.41e-239 | 312 | 2046 | 264 | 1928 | Callose synthase 7 OS=Arabidopsis thaliana OX=3702 GN=CALS7 PE=3 SV=3 |
|
| 1.05e-232 | 308 | 2047 | 241 | 1955 | Callose synthase 3 OS=Arabidopsis thaliana OX=3702 GN=CALS3 PE=3 SV=3 |
|
| 4.61e-230 | 308 | 2039 | 237 | 1937 | Callose synthase 2 OS=Arabidopsis thaliana OX=3702 GN=CALS2 PE=2 SV=3 |
|
| 1.57e-227 | 312 | 2045 | 252 | 1864 | Callose synthase 4 OS=Arabidopsis thaliana OX=3702 GN=CALS4 PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000038 | 0.000003 |
TMHMM Annotations download full data without filtering help
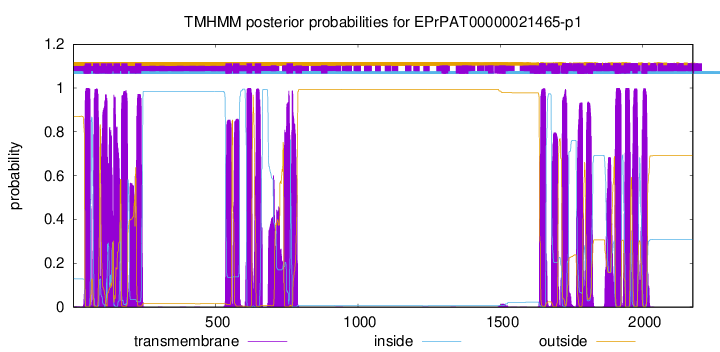
| Start | End |
|---|---|
| 41 | 63 |
| 70 | 92 |
| 97 | 116 |
| 118 | 137 |
| 142 | 164 |
| 169 | 191 |
| 217 | 239 |
| 535 | 557 |
| 561 | 583 |
| 609 | 628 |
| 638 | 660 |
| 749 | 771 |
| 1637 | 1659 |
| 1680 | 1702 |
| 1717 | 1739 |
| 1770 | 1792 |
| 1802 | 1824 |
| 1902 | 1924 |
| 1934 | 1956 |
| 1963 | 1982 |
| 1997 | 2016 |
