You are browsing environment: FUNGIDB
CAZyme Information: EPrPAT00000017534-p1
You are here: Home > Sequence: EPrPAT00000017534-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Pythium aphanidermatum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Pythiaceae; Pythium; Pythium aphanidermatum | |||||||||||
| CAZyme ID | EPrPAT00000017534-p1 | |||||||||||
| CAZy Family | GH17 | |||||||||||
| CAZyme Description | Alpha-glucosidase. | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.20:35 | 3.2.1.177:14 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH31 | 331 | 688 | 1.3e-103 | 0.6838407494145199 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 269888 | GH31_MGAM_SI_GAA | 8.36e-139 | 335 | 595 | 124 | 367 | maltase-glucoamylase, sucrase-isomaltase, lysosomal acid alpha-glucosidase. This subgroup includes the following three closely related glycosyl hydrolase family 31 (GH31) enzymes: maltase-glucoamylase (MGAM), sucrase-isomaltase (SI), and lysosomal acid alpha-glucosidase (GAA), also known as acid-maltase. MGAM is one of the two enzymes responsible for catalyzing the last glucose-releasing step in starch digestion. SI is implicated in the digestion of dietary starch and major disaccharides such as sucrose and isomaltose, while GAA degrades glycogen in the lysosome, cleaving both alpha-1,4 and alpha-1,6 glucosidic linkages. MGAM and SI are anchored to small-intestinal brush-border epithelial cells. The absence of SI from the brush border membrane or its malfunction is associated with malabsorption disorders such as congenital sucrase-isomaltase deficiency (CSID). The domain architectures of MGAM and SI include two tandem GH31 catalytic domains, an N-terminal domain found near the membrane-bound end, and a C-terminal luminal domain. Both of the tandem GH31 domains of MGAM and SI are included in this family. The domain architecture of GAA includes an N-terminal TFF (trefoil factor family) domain in addition to the GH31 catalytic domain. Deficient GAA expression causes Pompe disease, an autosomal recessive genetic disorder also known as glycogen storage disease type II (GSDII). |
| 395838 | Glyco_hydro_31 | 4.82e-136 | 324 | 688 | 133 | 442 | Glycosyl hydrolases family 31. Glycosyl hydrolases are key enzymes of carbohydrate metabolism. Family 31 comprises of enzymes that are, or similar to, alpha- galactosidases. |
| 269889 | GH31_GANC_GANAB_alpha | 2.96e-92 | 335 | 727 | 122 | 466 | neutral alpha-glucosidase C, neutral alpha-glucosidase AB. This subgroup includes the closely related glycosyl hydrolase family 31 (GH31) isozymes, neutral alpha-glucosidase C (GANC) and the alpha subunit of heterodimeric neutral alpha-glucosidase AB (GANAB). Initially distinguished on the basis of differences in electrophoretic mobility in starch gel, GANC and GANAB have been shown to have other differences, including those of substrate specificity. GANC and GANAB are key enzymes in glycogen metabolism that hydrolyze terminal, non-reducing 1,4-linked alpha-D-glucose residues from glycogen in the endoplasmic reticulum. The GANC/GANAB family includes the alpha-glucosidase II (ModA) from Dictyostelium discoideum as well as the alpha-glucosidase II (GLS2, or ROT2 - Reversal of TOR2 lethality protein 2) from Saccharomyces cerevisiae. |
| 224418 | YicI | 2.33e-84 | 331 | 759 | 373 | 740 | Alpha-glucosidase, glycosyl hydrolase family GH31 [Carbohydrate transport and metabolism]. |
| 269890 | GH31_glucosidase_II_MalA | 7.45e-68 | 335 | 598 | 122 | 339 | Alpha-glucosidase II-like. Alpha-glucosidase II (alpha-D-glucoside glucohydrolase) is a glycosyl hydrolase family 31 (GH31) enzyme, found in bacteria and plants, which has exo-alpha-1,4-glucosidase and oligo-1,6-glucosidase activities. Alpha-glucosidase II has been characterized in Bacillus thermoamyloliquefaciens where it forms a homohexamer. This subgroup also includes the MalA alpha-glucosidase from Sulfolobus solfataricus and the AglA alpha-glucosidase from Picrophilus torridus. MalA is part of the carbohydrate-metabolizing machinery that allows this organism to utilize carbohydrates, such as maltose, as the sole carbon and energy source. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 8.28e-144 | 174 | 726 | 57 | 789 | |
| 2.10e-141 | 177 | 815 | 54 | 888 | |
| 3.01e-140 | 170 | 781 | 51 | 852 | |
| 8.08e-137 | 177 | 744 | 57 | 815 | |
| 8.15e-134 | 178 | 806 | 61 | 891 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.76e-117 | 140 | 780 | 39 | 854 | Sugar beet alpha-glucosidase with acarbose [Beta vulgaris],3W38_A Sugar beet alpha-glucosidase [Beta vulgaris],3WEL_A Sugar beet alpha-glucosidase with acarviosyl-maltotriose [Beta vulgaris],3WEM_A Sugar beet alpha-glucosidase with acarviosyl-maltotetraose [Beta vulgaris],3WEN_A Sugar beet alpha-glucosidase with acarviosyl-maltopentaose [Beta vulgaris],3WEO_A Sugar beet alpha-glucosidase with acarviosyl-maltohexaose [Beta vulgaris] |
|
| 8.35e-96 | 335 | 806 | 403 | 857 | Crystal structure of human lysosomal acid-alpha-glucosidase, GAA [Homo sapiens],5NN5_A Crystal structure of human lysosomal acid-alpha-glucosidase, GAA, in complex with 1-deoxynojirimycin [Homo sapiens],5NN6_A Crystal structure of human lysosomal acid-alpha-glucosidase, GAA, in complex with N-hydroxyethyl-1-deoxynojirimycin [Homo sapiens],5NN8_A Crystal structure of human lysosomal acid-alpha-glucosidase, GAA, in complex with acarbose [Homo sapiens] |
|
| 8.35e-96 | 335 | 806 | 403 | 857 | Crystal structure of human lysosomal acid-alpha-glucosidase, GAA, in complex with N-acetyl-cysteine [Homo sapiens] |
|
| 8.68e-96 | 335 | 806 | 405 | 859 | Crystal structure of human GAA [Homo sapiens],5KZX_A Crystal structure of human GAA [Homo sapiens] |
|
| 3.57e-92 | 144 | 821 | 75 | 898 | Crystal structure of the N-terminal domain of sucrase-isomaltase [Homo sapiens],3LPO_B Crystal structure of the N-terminal domain of sucrase-isomaltase [Homo sapiens],3LPO_C Crystal structure of the N-terminal domain of sucrase-isomaltase [Homo sapiens],3LPO_D Crystal structure of the N-terminal domain of sucrase-isomaltase [Homo sapiens],3LPP_A Crystal complex of N-terminal sucrase-isomaltase with kotalanol [Homo sapiens],3LPP_B Crystal complex of N-terminal sucrase-isomaltase with kotalanol [Homo sapiens],3LPP_C Crystal complex of N-terminal sucrase-isomaltase with kotalanol [Homo sapiens],3LPP_D Crystal complex of N-terminal sucrase-isomaltase with kotalanol [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 8.80e-125 | 133 | 779 | 25 | 827 | Probable alpha-glucosidase Os06g0675700 OS=Oryza sativa subsp. japonica OX=39947 GN=Os06g0675700 PE=1 SV=1 |
|
| 6.87e-124 | 169 | 778 | 52 | 850 | Alpha-xylosidase 1 OS=Arabidopsis thaliana OX=3702 GN=XYL1 PE=1 SV=1 |
|
| 2.95e-123 | 181 | 776 | 59 | 817 | Alpha-glucosidase OS=Hordeum vulgare OX=4513 PE=2 SV=1 |
|
| 2.41e-119 | 132 | 778 | 26 | 848 | Alpha-glucosidase OS=Spinacia oleracea OX=3562 PE=1 SV=1 |
|
| 2.08e-111 | 140 | 780 | 39 | 854 | Alpha-glucosidase OS=Beta vulgaris OX=161934 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
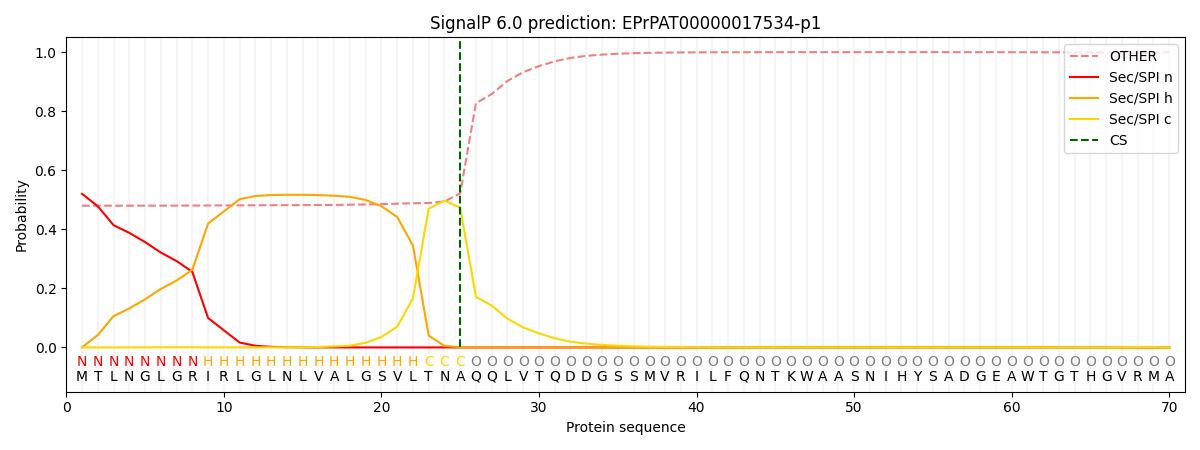
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.493186 | 0.506807 | CS pos: 25-26. Pr: 0.4733 |
