You are browsing environment: FUNGIDB
CAZyme Information: EPrPAT00000017417-p1
You are here: Home > Sequence: EPrPAT00000017417-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Pythium aphanidermatum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Pythiaceae; Pythium; Pythium aphanidermatum | |||||||||||
| CAZyme ID | EPrPAT00000017417-p1 | |||||||||||
| CAZy Family | GH16|GH16 | |||||||||||
| CAZyme Description | GlcNac transferase. | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT60 | 65 | 379 | 2.6e-58 | 0.996969696969697 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 371506 | GlcNAc | 3.79e-46 | 65 | 379 | 1 | 350 | Glycosyltransferase (GlcNAc). GlcNAc is an enzyme that carries out the first glycosylation step of hydroxylated Skp1, a ubiquitous eukaryotic protein, in the cytoplasm. |
| 397897 | Sas10_Utp3 | 5.14e-09 | 761 | 842 | 1 | 84 | Sas10/Utp3/C1D family. This family contains Utp3 and LCP5 which are components of the U3 ribonucleoprotein complex. It also includes the human C1D protein and Saccharomyces cerevisiae YHR081W (rrp47), an exosome-associated protein required for the 3' processing of stable RNAs, and Sas10 which has been identified as a regulator of chromatin silencing. This family also includes the human protein Neuroguidin an initiation factor 4E (eIF4E) binding protein. |
| 173412 | PTZ00121 | 4.27e-04 | 830 | 930 | 1404 | 1504 | MAEBL; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.87e-198 | 23 | 503 | 28 | 496 | |
| 1.10e-190 | 19 | 413 | 21 | 414 | |
| 8.37e-188 | 29 | 413 | 13 | 398 | |
| 8.01e-150 | 6 | 413 | 10 | 420 | |
| 2.58e-35 | 70 | 396 | 91 | 412 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.17e-15 | 762 | 870 | 19 | 122 | Nuclear nucleic acid-binding protein C1D OS=Gallus gallus OX=9031 GN=C1D PE=2 SV=1 |
|
| 1.55e-15 | 759 | 870 | 16 | 122 | Nuclear nucleic acid-binding protein C1D OS=Mus musculus OX=10090 GN=C1d PE=1 SV=1 |
|
| 2.11e-15 | 759 | 870 | 16 | 122 | Nuclear nucleic acid-binding protein C1D OS=Cricetulus griseus OX=10029 GN=C1D PE=2 SV=1 |
|
| 9.93e-15 | 759 | 870 | 16 | 122 | Nuclear nucleic acid-binding protein C1D OS=Homo sapiens OX=9606 GN=C1D PE=1 SV=1 |
|
| 9.93e-15 | 759 | 870 | 16 | 122 | Nuclear nucleic acid-binding protein C1D OS=Bos taurus OX=9913 GN=C1D PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
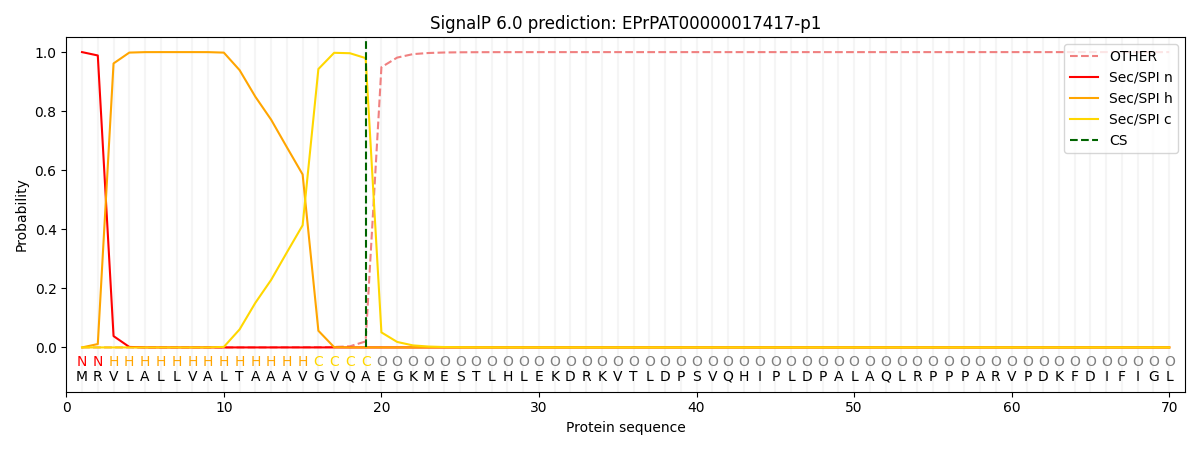
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000206 | 0.999763 | CS pos: 19-20. Pr: 0.9796 |

