You are browsing environment: FUNGIDB
CAZyme Information: EPrPAT00000015312-p1
You are here: Home > Sequence: EPrPAT00000015312-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
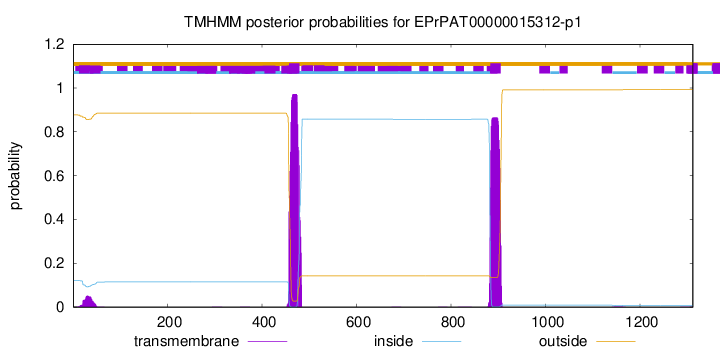
TMHMM annotations
Basic Information help
| Species | Pythium aphanidermatum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Pythiaceae; Pythium; Pythium aphanidermatum | |||||||||||
| CAZyme ID | EPrPAT00000015312-p1 | |||||||||||
| CAZy Family | AA4 | |||||||||||
| CAZyme Description | Cysteine protease family C54. | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT62 | 569 | 800 | 1.4e-43 | 0.9253731343283582 |
| GT62 | 1038 | 1268 | 1.3e-42 | 0.9216417910447762 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397471 | Peptidase_C54 | 1.86e-113 | 78 | 384 | 2 | 271 | Peptidase family C54. |
| 397491 | Anp1 | 4.44e-47 | 556 | 798 | 3 | 261 | Anp1. The members of this family (Anp1, Van1 and Mnn9) are membrane proteins required for proper Golgi function. These proteins co-localize within the cis Golgi, and that they are physically associated in two distinct complexes. |
| 397491 | Anp1 | 2.20e-43 | 1064 | 1266 | 37 | 261 | Anp1. The members of this family (Anp1, Van1 and Mnn9) are membrane proteins required for proper Golgi function. These proteins co-localize within the cis Golgi, and that they are physically associated in two distinct complexes. |
| 224137 | GT2 | 1.02e-04 | 673 | 842 | 70 | 219 | Glycosyltransferase, GT2 family [Carbohydrate transport and metabolism]. |
| 224137 | GT2 | 0.003 | 1142 | 1310 | 70 | 219 | Glycosyltransferase, GT2 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.47e-42 | 569 | 845 | 75 | 385 | |
| 3.47e-42 | 569 | 845 | 75 | 385 | |
| 1.89e-41 | 63 | 388 | 376 | 726 | |
| 5.70e-41 | 1035 | 1310 | 82 | 385 | |
| 1.05e-40 | 1040 | 1310 | 82 | 375 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 9.32e-59 | 56 | 407 | 23 | 361 | The crystal structure of human Atg4B [Homo sapiens] |
|
| 9.83e-59 | 56 | 407 | 25 | 363 | Structure of human Atg4b [Homo sapiens],2D1I_B Structure of human Atg4b [Homo sapiens] |
|
| 1.49e-56 | 56 | 403 | 23 | 357 | Chain A, Cysteine protease ATG4B [Homo sapiens],2Z0E_A Chain A, Cysteine protease ATG4B [Homo sapiens] |
|
| 1.49e-56 | 56 | 403 | 23 | 357 | Chain A, Cysteine protease ATG4B [Homo sapiens] |
|
| 4.71e-52 | 47 | 378 | 10 | 326 | Cysteine protease ATG4A [Homo sapiens],2P82_B Cysteine protease ATG4A [Homo sapiens],2P82_C Cysteine protease ATG4A [Homo sapiens],2P82_D Cysteine protease ATG4A [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 8.26e-60 | 56 | 407 | 20 | 358 | Cysteine protease ATG4B OS=Mus musculus OX=10090 GN=Atg4b PE=1 SV=2 |
|
| 4.07e-59 | 56 | 405 | 20 | 347 | Cysteine protease ATG4B OS=Xenopus laevis OX=8355 GN=atg4b PE=2 SV=1 |
|
| 7.05e-59 | 56 | 407 | 20 | 358 | Cysteine protease ATG4B OS=Rattus norvegicus OX=10116 GN=Atg4b PE=1 SV=1 |
|
| 1.11e-57 | 56 | 407 | 20 | 358 | Cysteine protease ATG4B OS=Homo sapiens OX=9606 GN=ATG4B PE=1 SV=2 |
|
| 1.51e-57 | 56 | 407 | 19 | 358 | Cysteine protease ATG4B OS=Gallus gallus OX=9031 GN=ATG4B PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
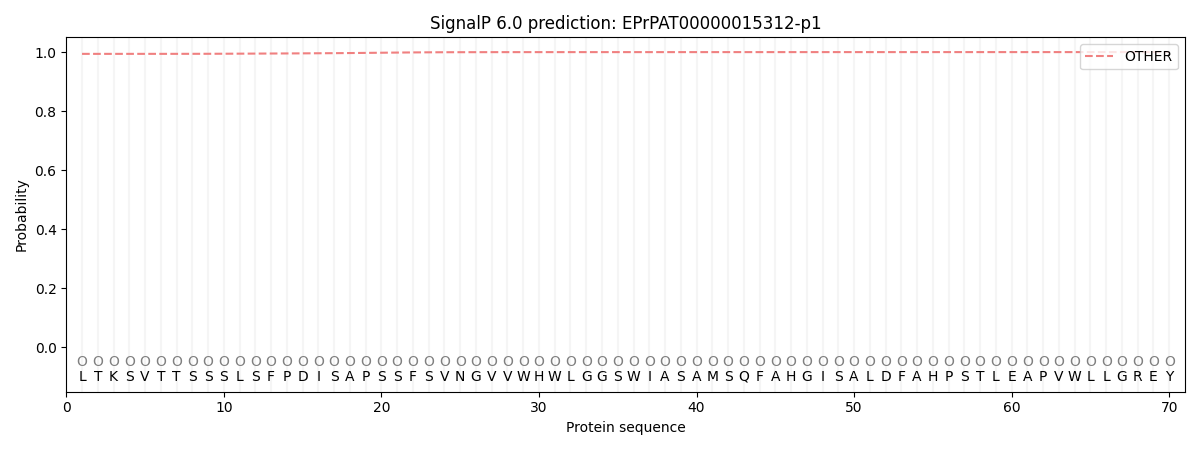
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.994852 | 0.005169 |