You are browsing environment: FUNGIDB
CAZyme Information: EPrPAT00000015033-p1
You are here: Home > Sequence: EPrPAT00000015033-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Pythium aphanidermatum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Pythiaceae; Pythium; Pythium aphanidermatum | |||||||||||
| CAZyme ID | EPrPAT00000015033-p1 | |||||||||||
| CAZy Family | AA2 | |||||||||||
| CAZyme Description | Callose synthase. | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.34:28 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT48 | 858 | 1593 | 2e-252 | 0.93234100135318 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396784 | Glucan_synthase | 9.65e-162 | 857 | 1575 | 2 | 716 | 1,3-beta-glucan synthase component. This family consists of various 1,3-beta-glucan synthase components including Gls1, Gls2 and Gls3 from yeast. 1,3-beta-glucan synthase EC:2.4.1.34 also known as callose synthase catalyzes the formation of a beta-1,3-glucan polymer that is a major component of the fungal cell wall. The reaction catalyzed is:- UDP-glucose + {(1,3)-beta-D-glucosyl}(N) <=> UDP + {(1,3)-beta-D-glucosyl}(N+1). |
| 405046 | FKS1_dom1 | 4.94e-33 | 137 | 232 | 5 | 106 | 1,3-beta-glucan synthase subunit FKS1, domain-1. The FKS1_dom1 domain is likely to be the 'Class I' region just N-terminal to the first set of transmembrane helices that is involved in 1,3-beta-glucan synthesis itself. This family is found on proteins with family Glucan_synthase, pfam02364. |
| 395036 | Sugar_tr | 4.49e-14 | 1774 | 2203 | 2 | 434 | Sugar (and other) transporter. |
| 273317 | SP | 2.30e-09 | 1843 | 2203 | 85 | 467 | MFS transporter, sugar porter (SP) family. This model represent the sugar porter subfamily of the major facilitator superfamily (pfam00083) [Transport and binding proteins, Carbohydrates, organic alcohols, and acids] |
| 340915 | MFS_GLUT_Class1_2_like | 3.06e-08 | 1774 | 2203 | 1 | 438 | Class 1 and Class 2 Glucose transporters (GLUTs) of the Major Facilitator Superfamily. This subfamily includes Class 1 and Class 2 glucose transporters (GLUTs) including Solute carrier family 2, facilitated glucose transporter member 1 (SLC2A1, also called glucose transporter type 1 or GLUT1), SLC2A2-5 (GLUT2-5), SLC2A7 (GLUT7), SLC2A9 (GLUT9), SLC2A11 (GLUT11), SLC2A14 (GLUT14), and similar proteins. GLUTs are a family of proteins that facilitate the transport of hexoses such as glucose and fructose. There are fourteen GLUTs found in humans; they display different substrate specificities and tissue expression. They have been categorized into three classes based on sequence similarity: Class 1 (GLUTs 1-4, 14); Class 2 (GLUTs 5, 7, 9, and 11); and Class 3 (GLUTs 6, 8, 10, 12, and HMIT). GLUTs 1-5 are the most thoroughly studied and are well-established as glucose and/or fructose transporters in various tissues and cell types. GLUT proteins are comprised of about 500 amino acid residues, possess a single N-linked oligosaccharide, and have 12 transmembrane segments. They belong to the Glucose transporter -like (GLUT-like) family of the Major Facilitator Superfamily (MFS) of membrane transport proteins. MFS proteins are thought to function through a single substrate binding site, alternating-access mechanism involving a rocker-switch type of movement. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 12 | 2417 | 12 | 2450 | |
| 0.0 | 42 | 2253 | 82 | 2263 | |
| 0.0 | 55 | 2220 | 43 | 2255 | |
| 0.0 | 55 | 2235 | 55 | 2228 | |
| 0.0 | 57 | 2121 | 92 | 2081 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.82e-209 | 55 | 1763 | 237 | 1936 | Callose synthase 3 OS=Arabidopsis thaliana OX=3702 GN=CALS3 PE=3 SV=3 |
|
| 5.82e-205 | 55 | 1763 | 233 | 1931 | Callose synthase 2 OS=Arabidopsis thaliana OX=3702 GN=CALS2 PE=2 SV=3 |
|
| 2.83e-204 | 55 | 1763 | 233 | 1931 | Callose synthase 1 OS=Arabidopsis thaliana OX=3702 GN=CALS1 PE=1 SV=2 |
|
| 1.15e-203 | 55 | 1763 | 240 | 1910 | Callose synthase 5 OS=Arabidopsis thaliana OX=3702 GN=CALS5 PE=1 SV=1 |
|
| 2.19e-203 | 55 | 1770 | 256 | 1922 | Callose synthase 7 OS=Arabidopsis thaliana OX=3702 GN=CALS7 PE=3 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
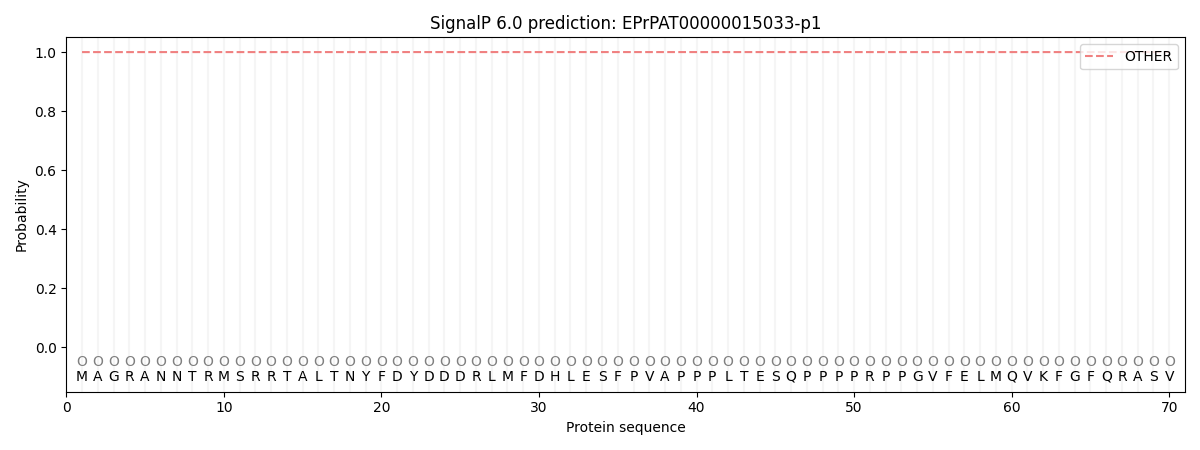
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000060 | 0.000000 |
TMHMM Annotations download full data without filtering help
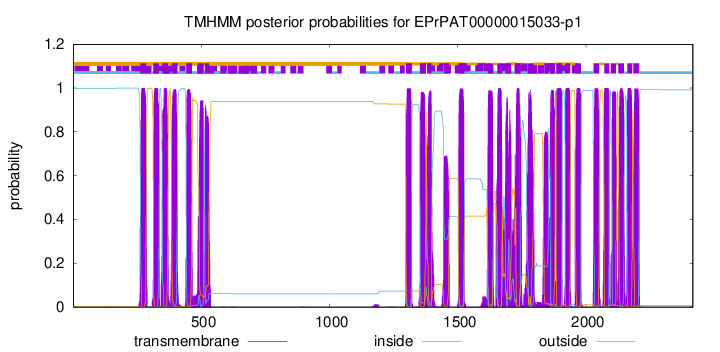
| Start | End |
|---|---|
| 263 | 285 |
| 312 | 334 |
| 347 | 369 |
| 384 | 406 |
| 441 | 463 |
| 486 | 508 |
| 513 | 530 |
| 1298 | 1320 |
| 1353 | 1375 |
| 1380 | 1397 |
| 1445 | 1467 |
| 1503 | 1525 |
| 1617 | 1639 |
| 1654 | 1673 |
| 1686 | 1708 |
| 1723 | 1745 |
| 1773 | 1795 |
| 1837 | 1854 |
| 1861 | 1879 |
| 1884 | 1906 |
| 1919 | 1938 |
| 1958 | 1980 |
| 2029 | 2051 |
| 2071 | 2090 |
| 2097 | 2116 |
| 2126 | 2148 |
| 2161 | 2180 |
| 2185 | 2207 |
