You are browsing environment: FUNGIDB
CAZyme Information: EPrPAT00000013567-p1
You are here: Home > Sequence: EPrPAT00000013567-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Pythium aphanidermatum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Pythiaceae; Pythium; Pythium aphanidermatum | |||||||||||
| CAZyme ID | EPrPAT00000013567-p1 | |||||||||||
| CAZy Family | AA17 | |||||||||||
| CAZyme Description | Transmembrane protein. | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA7 | 41 | 478 | 7.3e-89 | 0.980349344978166 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 223354 | GlcD | 1.05e-29 | 42 | 480 | 27 | 458 | FAD/FMN-containing dehydrogenase [Energy production and conversion]. |
| 396238 | FAD_binding_4 | 1.82e-22 | 47 | 188 | 1 | 139 | FAD binding domain. This family consists of various enzymes that use FAD as a co-factor, most of the enzymes are similar to oxygen oxidoreductase. One of the enzymes Vanillyl-alcohol oxidase (VAO) has a solved structure, the alignment includes the FAD binding site, called the PP-loop, between residues 99-110. The FAD molecule is covalently bound in the known structure, however the residue that links to the FAD is not in the alignment. VAO catalyzes the oxidation of a wide variety of substrates, ranging form aromatic amines to 4-alkylphenols. Other members of this family include D-lactate dehydrogenase, this enzyme catalyzes the conversion of D-lactate to pyruvate using FAD as a co-factor; mitomycin radical oxidase, this enzyme oxidizes the reduced form of mitomycins and is involved in mitomycin resistance. This family includes MurB an UDP-N-acetylenolpyruvoylglucosamine reductase enzyme EC:1.1.1.158. This enzyme is involved in the biosynthesis of peptidoglycan. |
| 369658 | BBE | 1.11e-16 | 433 | 479 | 1 | 45 | Berberine and berberine like. This domain is found in the berberine bridge and berberine bridge- like enzymes which are involved in the biosynthesis of numerous isoquinoline alkaloids. They catalyze the transformation of the N-methyl group of (S)-reticuline into the C-8 berberine bridge carbon of (S)-scoulerine. |
| 341037 | MFS_FBT | 1.74e-13 | 628 | 896 | 44 | 295 | Folate-biopterin transporter of the Major Facilitator Superfamily of transporters. The Folate-biopterin transporter (FBT) family includes folate carriers related to those of trypanosomatids in higher plant plastids and cyanobacteria. FBT mediates folate monoglutamate transport involved in tetrahydrofolate biosynthesis. It also mediates transport of antifolates, such as methotrexate and aminopterin. The FBT family belongs to the Major Facilitator Superfamily (MFS) of membrane transport proteins, which are thought to function through a single substrate binding site, alternating-access mechanism involving a rocker-switch type of movement. |
| 341037 | MFS_FBT | 2.22e-12 | 1170 | 1429 | 45 | 286 | Folate-biopterin transporter of the Major Facilitator Superfamily of transporters. The Folate-biopterin transporter (FBT) family includes folate carriers related to those of trypanosomatids in higher plant plastids and cyanobacteria. FBT mediates folate monoglutamate transport involved in tetrahydrofolate biosynthesis. It also mediates transport of antifolates, such as methotrexate and aminopterin. The FBT family belongs to the Major Facilitator Superfamily (MFS) of membrane transport proteins, which are thought to function through a single substrate binding site, alternating-access mechanism involving a rocker-switch type of movement. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.49e-45 | 22 | 478 | 12 | 470 | |
| 2.02e-44 | 22 | 478 | 37 | 495 | |
| 8.04e-44 | 26 | 480 | 45 | 492 | |
| 1.28e-43 | 7 | 482 | 17 | 486 | |
| 1.44e-43 | 46 | 480 | 64 | 491 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.16e-46 | 4 | 479 | 55 | 528 | AtBBE15 [Arabidopsis thaliana],4UD8_B AtBBE15 [Arabidopsis thaliana] |
|
| 3.07e-46 | 8 | 478 | 4 | 476 | The crystal structure of an Acremonium strictum glucooligosaccharide oxidase reveals a novel flavinylation [Sarocladium strictum],2AXR_A Crystal structure of glucooligosaccharide oxidase from Acremonium strictum: a novel flavinylation of 6-S-cysteinyl, 8alpha-N1-histidyl FAD [Sarocladium strictum] |
|
| 4.54e-45 | 8 | 478 | 4 | 476 | Chain A, Glucooligosaccharide oxidase [Sarocladium strictum] |
|
| 7.89e-45 | 5 | 480 | 52 | 528 | Structure of BBE-like #28 from Arabidopsis thaliana [Arabidopsis thaliana],5D79_B Structure of BBE-like #28 from Arabidopsis thaliana [Arabidopsis thaliana] |
|
| 2.32e-44 | 29 | 480 | 30 | 470 | Physcomitrella patens BBE-like 1 wild-type [Physcomitrium patens],6EO4_B Physcomitrella patens BBE-like 1 wild-type [Physcomitrium patens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 9.31e-53 | 32 | 485 | 60 | 540 | FAD-linked oxidoreductase pynB OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=pynB PE=1 SV=1 |
|
| 3.09e-50 | 30 | 479 | 16 | 444 | Uncharacterized FAD-linked oxidoreductase YvdP OS=Bacillus subtilis (strain 168) OX=224308 GN=yvdP PE=1 SV=1 |
|
| 5.17e-49 | 26 | 479 | 12 | 443 | Uncharacterized FAD-linked oxidoreductase YgaK OS=Bacillus subtilis (strain 168) OX=224308 GN=ygaK PE=3 SV=4 |
|
| 7.18e-48 | 46 | 479 | 83 | 530 | Berberine bridge enzyme-like 13 OS=Arabidopsis thaliana OX=3702 GN=At1g30760 PE=1 SV=1 |
|
| 3.52e-47 | 10 | 479 | 65 | 533 | Berberine bridge enzyme-like 21 OS=Arabidopsis thaliana OX=3702 GN=At4g20840 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
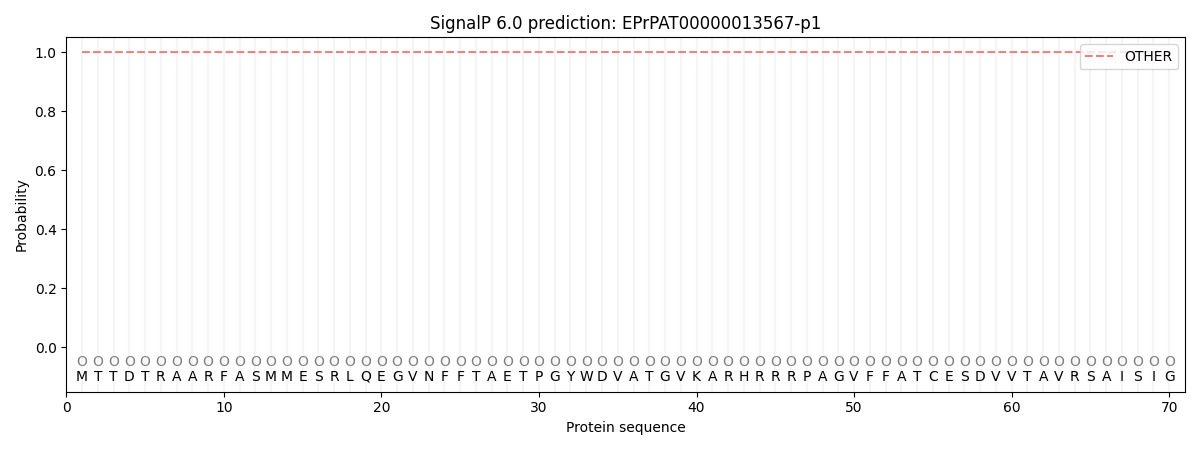
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000059 | 0.000000 |
TMHMM Annotations download full data without filtering help
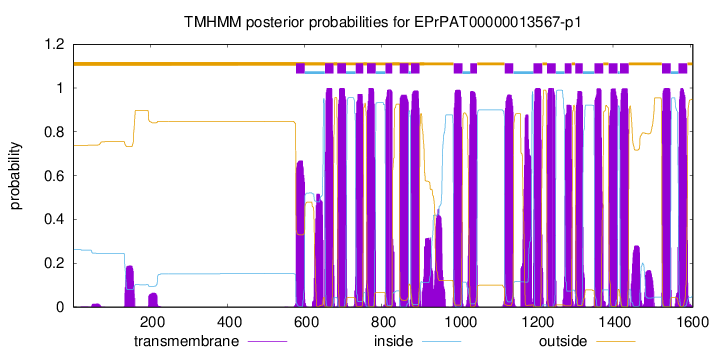
| Start | End |
|---|---|
| 578 | 600 |
| 653 | 675 |
| 685 | 707 |
| 733 | 752 |
| 762 | 784 |
| 810 | 827 |
| 847 | 869 |
| 876 | 898 |
| 987 | 1009 |
| 1030 | 1047 |
| 1119 | 1141 |
| 1194 | 1216 |
| 1229 | 1251 |
| 1275 | 1292 |
| 1302 | 1321 |
| 1352 | 1374 |
| 1389 | 1411 |
| 1418 | 1440 |
| 1527 | 1549 |
| 1570 | 1592 |
