You are browsing environment: FUNGIDB
CAZyme Information: EPE08465.1
You are here: Home > Sequence: EPE08465.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Ophiostoma piceae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Ophiostomataceae; Ophiostoma; Ophiostoma piceae | |||||||||||
| CAZyme ID | EPE08465.1 | |||||||||||
| CAZy Family | GH76 | |||||||||||
| CAZyme Description | Peroxidase [Source:UniProtKB/TrEMBL;Acc:S3D4X9] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA2 | 65 | 206 | 1.8e-26 | 0.5686274509803921 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 173829 | plant_peroxidase_like_1 | 1.02e-127 | 25 | 288 | 1 | 264 | Uncharacterized family of plant peroxidase-like proteins. This is a subgroup of heme-dependent peroxidases similar to plant peroxidases. Along with animal peroxidases, these enzymes belong to a group of peroxidases containing a heme prosthetic group (ferriprotoporphyrin IX) which catalyzes a multistep oxidative reaction involving hydrogen peroxide as the electron acceptor. The plant peroxidase-like superfamily is found in all three kingdoms of life and carries out a variety of biosynthetic and degradative functions. |
| 340867 | MFS_unc93-like | 1.28e-62 | 582 | 990 | 1 | 410 | Uncharacterized Unc-93-like proteins of the Major Facilitator Superfamily of transporters. This subfamily consists of uncharacterized proteins, mainly from fungi and plants, with similarity to Caenorhabditis elegans uncoordinated protein 93 (also called putative potassium channel regulatory protein unc-93). Unc-93 acts as a regulatory subunit of a multi-subunit potassium channel complex that may function in coordinating muscle contraction in C. elegans. The unc93-like subfamily belongs to the Unc-93 family of the Major Facilitator Superfamily (MFS) of transporters. MFS proteins are thought to function through a single substrate binding site, alternating-access mechanism involving a rocker-switch type of movement. |
| 395089 | peroxidase | 8.07e-23 | 59 | 199 | 9 | 155 | Peroxidase. |
| 173823 | plant_peroxidase_like | 1.59e-22 | 66 | 288 | 18 | 255 | Heme-dependent peroxidases similar to plant peroxidases. Along with animal peroxidases, these enzymes belong to a group of peroxidases containing a heme prosthetic group (ferriprotoporphyrin IX), which catalyzes a multistep oxidative reaction involving hydrogen peroxide as the electron acceptor. The plant peroxidase-like superfamily is found in all three kingdoms of life and carries out a variety of biosynthetic and degradative functions. Several sub-families can be identified. Class I includes intracellular peroxidases present in fungi, plants, archaea and bacteria, called catalase-peroxidases, that can exhibit both catalase and broad-spectrum peroxidase activities depending on the steady-state concentration of hydrogen peroxide. Catalase-peroxidases are typically comprised of two homologous domains that probably arose via a single gene duplication event. Class II includes ligninase and other extracellular fungal peroxidases, while class III is comprised of classic extracellular plant peroxidases, like horseradish peroxidase. |
| 340896 | MFS_unc93_like | 2.14e-17 | 589 | 997 | 3 | 387 | Unc-93 family of the Major Facilitator Superfamily of transporters. The Unc-93 family is composed of Caenorhabditis elegans uncoordinated protein 93 (also called putative potassium channel regulatory protein unc-93) and similar proteins including three vertebrate members: protein unc-93 homolog A (UNC93A), protein unc-93 homolog B1 (UNC93B1), and UNC93-like protein MFSD11 (also called major facilitator superfamily domain-containing protein 11 or protein ET). Unc-93 acts as a regulatory subunit of a multi-subunit potassium channel complex that may function in coordinating muscle contraction in C. elegans. The human UNC93A gene is located in a region of the genome that is frequently associated with ovarian cancer, however, there is no evidence that UNC93A has a tumor suppressor function. UNC93B1 controls intracellular trafficking and transport of a subset of Toll-like receptors (TLRs), including TLR3, TLR7 and TLR9, from the endoplasmic reticulum to endolysosomes where they can engage pathogen nucleotides and activate signaling cascades. MFSD11 is ubiquitously expressed in the periphery and the central nervous system of mice, where it is expressed in excitatory and inhibitory mouse brain neurons. The unc93-like family belongs to the Major Facilitator Superfamily (MFS) of membrane transport proteins, which are thought to function through a single substrate binding site, alternating-access mechanism involving a rocker-switch type of movement. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 9.04e-116 | 17 | 523 | 18 | 526 | |
| 7.95e-109 | 11 | 505 | 8 | 508 | |
| 1.71e-108 | 10 | 501 | 6 | 504 | |
| 1.82e-103 | 5 | 527 | 9 | 529 | |
| 2.87e-97 | 24 | 497 | 21 | 497 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.53e-10 | 68 | 202 | 40 | 178 | The crystal structures of several mutants of pleurotus eryngii versatile peroxidase [Pleurotus eryngii] |
|
| 7.45e-10 | 68 | 243 | 41 | 223 | CRYSTAL STRUCTURE OF FUNGAL VERSATILE PEROXIDASE I FROM PLEUROTUS OSTREATUS - CRYSTAL FORM I [Pleurotus ostreatus],4BLL_A CRYSTAL STRUCTURE OF FUNGAL VERSATILE PEROXIDASE I FROM PLEUROTUS OSTREATUS - CRYSTAL FORM II [Pleurotus ostreatus],4BLN_A CRYSTAL STRUCTURE OF FUNGAL VERSATILE PEROXIDASE I FROM PLEUROTUS OSTREATUS - CRYSTAL FORM III [Pleurotus ostreatus],4BLX_A CRYSTAL STRUCTURE OF FUNGAL VERSATILE PEROXIDASE I FROM PLEUROTUS OSTREATUS - CRYSTAL FORM IV [Pleurotus ostreatus],4BLY_A Crystal Structure Of Fungal Versatile Peroxidase I From Pleurotus Ostreatus - Crystal Form V [Pleurotus ostreatus],4BLZ_A Crystal Structure Of Fungal Versatile Peroxidase I From Pleurotus Ostreatus - Crystal Form Vi [Pleurotus ostreatus],4BLZ_B Crystal Structure Of Fungal Versatile Peroxidase I From Pleurotus Ostreatus - Crystal Form Vi [Pleurotus ostreatus],4BM0_A Crystal Structure Of Fungal Versatile Peroxidase I From Pleurotus Ostreatus - Crystal Form Vii [Pleurotus ostreatus] |
|
| 9.85e-10 | 70 | 202 | 43 | 180 | Chain A, lignin peroxidase [Agrocybe pediades] |
|
| 1.75e-09 | 68 | 243 | 40 | 222 | Chain A, Versatile peroxidase VPL2 [Pleurotus eryngii] |
|
| 2.33e-09 | 68 | 243 | 40 | 222 | Chain A, Versatile peroxidase VPL2 [Pleurotus eryngii] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.29e-101 | 23 | 408 | 21 | 408 | WSC domain-containing protein ARB_07870 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_07870 PE=1 SV=1 |
|
| 3.49e-63 | 569 | 966 | 4 | 398 | Notoamide biosynthesis cluster protein O' OS=Aspergillus versicolor OX=46472 GN=notO' PE=1 SV=1 |
|
| 5.18e-39 | 569 | 1008 | 23 | 456 | UNC93-like protein 1 OS=Arabidopsis thaliana OX=3702 GN=At1g18000 PE=2 SV=2 |
|
| 5.18e-39 | 569 | 1008 | 23 | 456 | UNC93-like protein 2 OS=Arabidopsis thaliana OX=3702 GN=At1g18010 PE=2 SV=2 |
|
| 2.20e-32 | 563 | 959 | 31 | 440 | UNC93-like protein C922.05c OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=SPAC922.05c PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000713 | 0.999254 | CS pos: 19-20. Pr: 0.9723 |
TMHMM Annotations download full data without filtering help
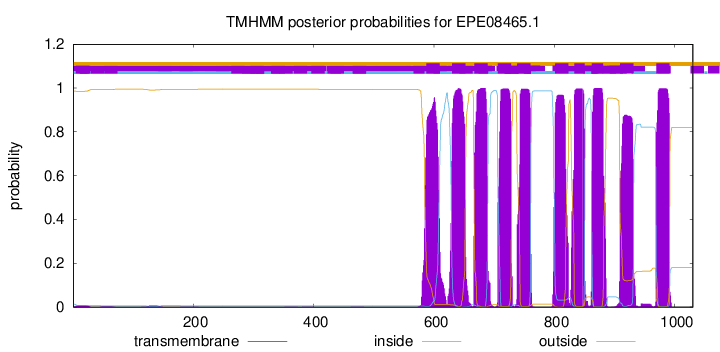
| Start | End |
|---|---|
| 587 | 609 |
| 630 | 652 |
| 667 | 689 |
| 706 | 728 |
| 738 | 760 |
| 801 | 823 |
| 833 | 850 |
| 862 | 881 |
| 909 | 931 |
| 969 | 991 |
