You are browsing environment: FUNGIDB
CAZyme Information: EPE02551.1
You are here: Home > Sequence: EPE02551.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Ophiostoma piceae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Ophiostomataceae; Ophiostoma; Ophiostoma piceae | |||||||||||
| CAZyme ID | EPE02551.1 | |||||||||||
| CAZy Family | AA11 | |||||||||||
| CAZyme Description | GH16 domain-containing protein [Source:UniProtKB/TrEMBL;Acc:S3BSZ5] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH16 | 55 | 328 | 3.7e-46 | 0.991304347826087 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 185691 | GH16_Strep_laminarinase_like | 9.09e-102 | 48 | 329 | 1 | 259 | Streptomyces laminarinase-like, member of glycosyl hydrolase family 16. Proteins similar to Streptomyces sioyaensis beta-1,3-glucanase (laminarinase) present in Actinomycetales as well as Peziomycotina. Laminarinases belong to glycosyl hydrolase family 16 and hydrolyze the glycosidic bond of the 1,3-beta-linked glucan, a major component of fungal and plant cell walls and the structural and storage polysaccharides (laminarin) of marine macro-algae. Members of the GH16 family have a conserved jelly roll fold with an active site channel. |
| 185693 | GH16_laminarinase_like | 4.03e-33 | 54 | 328 | 1 | 235 | Laminarinase, member of the glycosyl hydrolase family 16. Laminarinase, also known as glucan endo-1,3-beta-D-glucosidase, is a glycosyl hydrolase family 16 member that hydrolyzes 1,3-beta-D-glucosidic linkages in 1,3-beta-D-glucans such as laminarins, curdlans, paramylons, and pachymans, with very limited action on mixed-link (1,3-1,4-)-beta-D-glucans. |
| 185690 | GH16_fungal_Lam16A_glucanase | 1.65e-06 | 164 | 227 | 95 | 147 | fungal 1,3(4)-beta-D-glucanases, similar to Phanerochaete chrysosporium laminarinase 16A. Group of fungal 1,3(4)-beta-D-glucanases, similar to Phanerochaete chrysosporium laminarinase 16A. Lam16A belongs to the 'nonspecific' 1,3(4)-beta-glucanase subfamily, although beta-1,6 branching and beta-1,4 bonds specifically define where Lam16A hydrolyzes its substrates, like curdlan (beta-1,3-glucan), lichenin (beta-1,3-1,4-mixed linkage glucan), and laminarin (beta-1,6-branched-1,3-glucan). |
| 185694 | GH16_CCF | 5.20e-05 | 165 | 327 | 129 | 328 | Coelomic cytolytic factor, member of glycosyl hydrolase family 16. Subgroup of glucanases of unknown function that are related to beta-GRP (beta-1,3-glucan recognition protein), but contain active site residues. Beta-GRPs are one group of pattern recognition receptors (PRRs), also referred to as biosensor proteins, that complexes with pathogen-associated beta-1,3-glucans and then transduces signals necessary for activation of an appropriate innate immune response. Beta-GRPs are present in insects and lack all catalytic residues. This subgroup contains related proteins that still contain the active site and are widely distributed in eukaryotes. Their structures adopt a jelly roll fold with a deep active site channel harboring the catalytic residues, like those of other glycosyl hydrolase family 16 members. |
| 185689 | GH16_fungal_KRE6_glucanase | 0.007 | 86 | 328 | 30 | 294 | Saccharomyces cerevisiae KRE6 and related glucanses, member of glycosyl hydrolase family 16. KRE6 is a Saccharomyces cerevisiae glucanase that participates in the synthesis of beta-1,6-glucan, a major structural component of the cell wall. It is a golgi membrane protein required for normal beta-1,6-glucan levels in the cell wall. KRE6 is closely realted to laminarinase, a glycosyl hydrolase family 16 member that hydrolyzes 1,3-beta-D-glucosidic linkages in 1,3-beta-D-glucans such as laminarins, curdlans, paramylons, and pachymans, with very limited action on mixed-link (1,3-1,4-)-beta-D-glucans. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.52e-91 | 46 | 328 | 35 | 308 | |
| 2.26e-89 | 29 | 328 | 24 | 319 | |
| 2.11e-86 | 43 | 328 | 102 | 385 | |
| 4.21e-86 | 43 | 328 | 102 | 385 | |
| 5.03e-83 | 34 | 330 | 41 | 333 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.04e-60 | 46 | 327 | 6 | 273 | The 1.5 A crystal structure of endo-1,3-beta-glucanase from Streptomyces sioyaensis [Streptomyces sioyaensis] |
|
| 3.90e-58 | 51 | 330 | 11 | 280 | Chain A, 1,3-beta-glucanase [Actinomycetia bacterium] |
|
| 1.46e-20 | 53 | 328 | 6 | 238 | endo-1,3-beta-glucanase from Cellulosimicrobium cellulans [Cellulosimicrobium cellulans] |
|
| 9.00e-14 | 51 | 328 | 8 | 242 | The crystal structure of an endo-beta-1,3-glucanase from alkaliphilic Nocardiopsis sp.strain F96 [Nocardiopsis sp. F96] |
|
| 2.79e-11 | 63 | 329 | 23 | 260 | The X-ray structure of endo-beta-1,3-glucanase from Pyrococcus furiosus [Pyrococcus furiosus],2VY0_B The X-ray structure of endo-beta-1,3-glucanase from Pyrococcus furiosus [Pyrococcus furiosus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.87e-08 | 53 | 328 | 28 | 252 | Clotting factor G alpha subunit OS=Tachypleus tridentatus OX=6853 PE=1 SV=1 |
|
| 7.00e-07 | 65 | 298 | 53 | 259 | Beta-glucanase OS=Rhodothermus marinus OX=29549 GN=bglA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
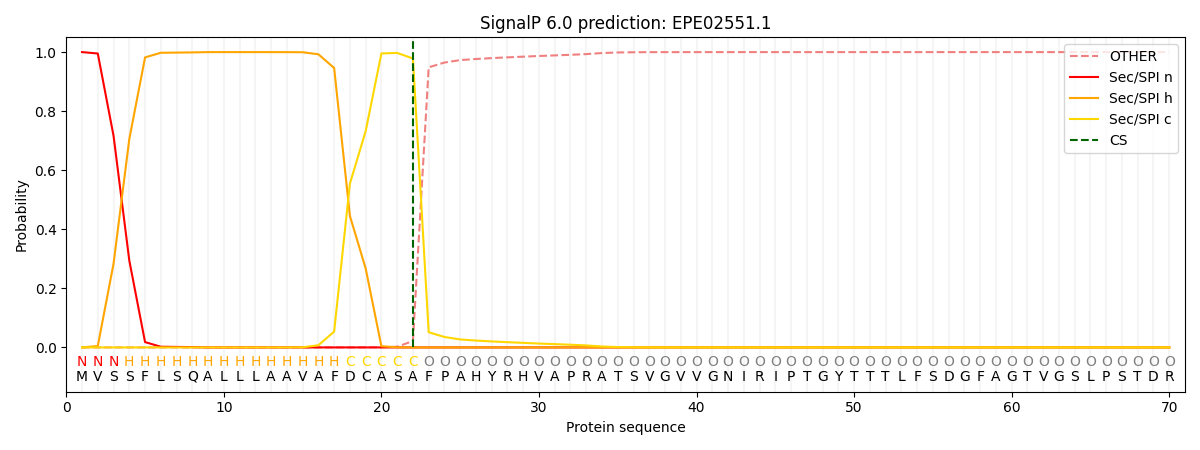
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000229 | 0.999753 | CS pos: 22-23. Pr: 0.9784 |
