You are browsing environment: FUNGIDB
CAZyme Information: ELR10480.1
You are here: Home > Sequence: ELR10480.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
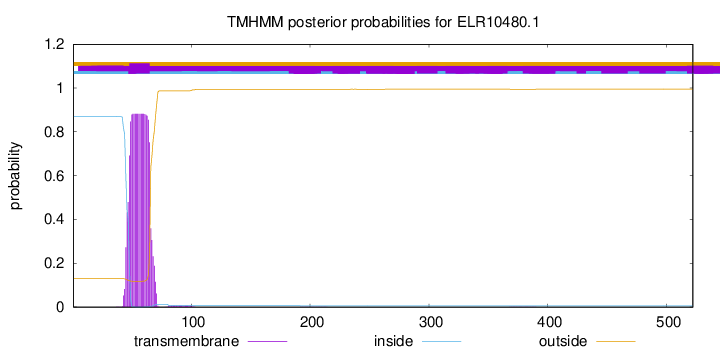
TMHMM annotations
Basic Information help
| Species | Pseudogymnoascus destructans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Leotiomycetes; ; Pseudeurotiaceae; Pseudogymnoascus; Pseudogymnoascus destructans | |||||||||||
| CAZyme ID | ELR10480.1 | |||||||||||
| CAZy Family | GT62 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT69 | 83 | 348 | 2.7e-68 | 0.9832635983263598 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 403053 | CAP59_mtransfer | 5.56e-89 | 83 | 344 | 2 | 225 | Cryptococcal mannosyltransferase 1. The capsule of pathogenic fungi is a complex polysaccharide whose formation is determined by a number of enzymes including, most importantly, alpha-1,3-mannosyltransferase 1, EC:2.4.1.-. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.16e-168 | 5 | 521 | 9 | 589 | |
| 2.46e-166 | 5 | 522 | 7 | 586 | |
| 1.17e-165 | 5 | 521 | 18 | 601 | |
| 2.12e-160 | 40 | 520 | 47 | 518 | |
| 1.14e-118 | 64 | 513 | 83 | 474 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as OTHER
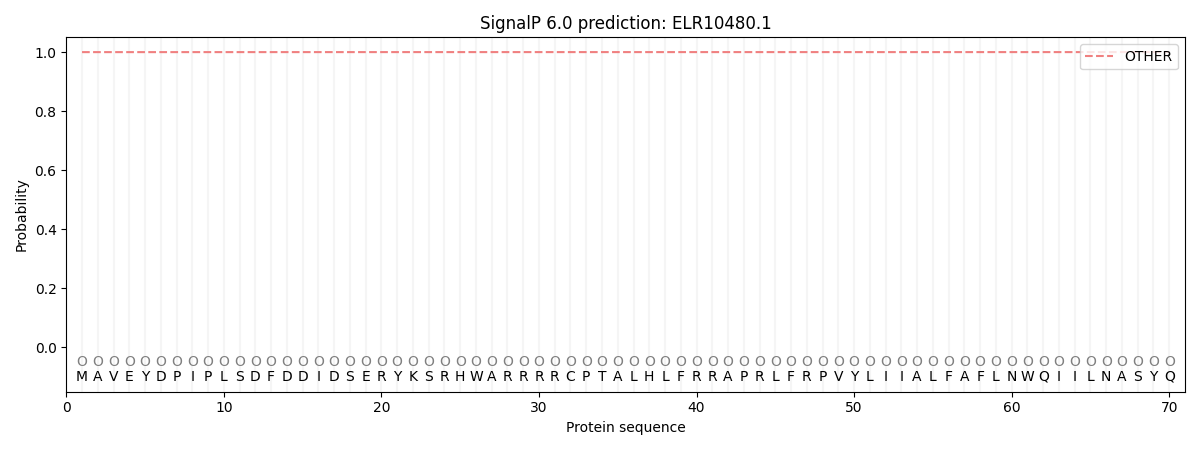
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999985 | 0.000020 |