You are browsing environment: FUNGIDB
CAZyme Information: ELR08699.1
You are here: Home > Sequence: ELR08699.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Pseudogymnoascus destructans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Leotiomycetes; ; Pseudeurotiaceae; Pseudogymnoascus; Pseudogymnoascus destructans | |||||||||||
| CAZyme ID | ELR08699.1 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.74e-35 | 40 | 201 | 141 | 287 | |
| 2.72e-32 | 45 | 201 | 147 | 310 | |
| 1.11e-28 | 40 | 199 | 160 | 309 | |
| 1.14e-28 | 45 | 201 | 150 | 314 | |
| 2.73e-28 | 40 | 203 | 145 | 304 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.81e-10 | 49 | 154 | 128 | 226 | Native structure of endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3PZG_A I222 crystal form of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3PZI_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 in complex with beta-D-glucose [Thermotoga petrophila RKU-1],3PZM_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with three glycerol molecules [Thermotoga petrophila RKU-1],3PZN_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with citrate and glycerol [Thermotoga petrophila RKU-1],3PZO_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 in complex with three maltose molecules [Thermotoga petrophila RKU-1],3PZQ_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with maltose and glycerol [Thermotoga petrophila RKU-1] |
|
| 5.82e-10 | 49 | 154 | 114 | 212 | X-ray structure of the endo-beta-1,4-mannanase from Thermotoga petrophila [Thermotoga petrophila RKU-1] |
|
| 1.95e-07 | 73 | 154 | 131 | 221 | Crystal Structure Analysis of the Endo-1,4-beta-mannanase from Rhizomucor miehei [Rhizomucor miehei] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.56e-11 | 49 | 154 | 131 | 237 | Mannan endo-1,4-beta-mannosidase 4 OS=Oryza sativa subsp. japonica OX=39947 GN=MAN4 PE=2 SV=2 |
|
| 4.62e-11 | 44 | 171 | 105 | 231 | Mannan endo-1,4-beta-mannosidase 8 OS=Oryza sativa subsp. japonica OX=39947 GN=MAN8 PE=2 SV=2 |
|
| 2.37e-10 | 48 | 154 | 153 | 260 | Mannan endo-1,4-beta-mannosidase 6 OS=Arabidopsis thaliana OX=3702 GN=MAN6 PE=2 SV=1 |
|
| 4.28e-10 | 38 | 173 | 139 | 299 | Mannan endo-1,4-beta-mannosidase 5 OS=Arabidopsis thaliana OX=3702 GN=MAN5 PE=2 SV=1 |
|
| 7.85e-10 | 38 | 174 | 140 | 296 | Mannan endo-1,4-beta-mannosidase 2 OS=Arabidopsis thaliana OX=3702 GN=MAN2 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
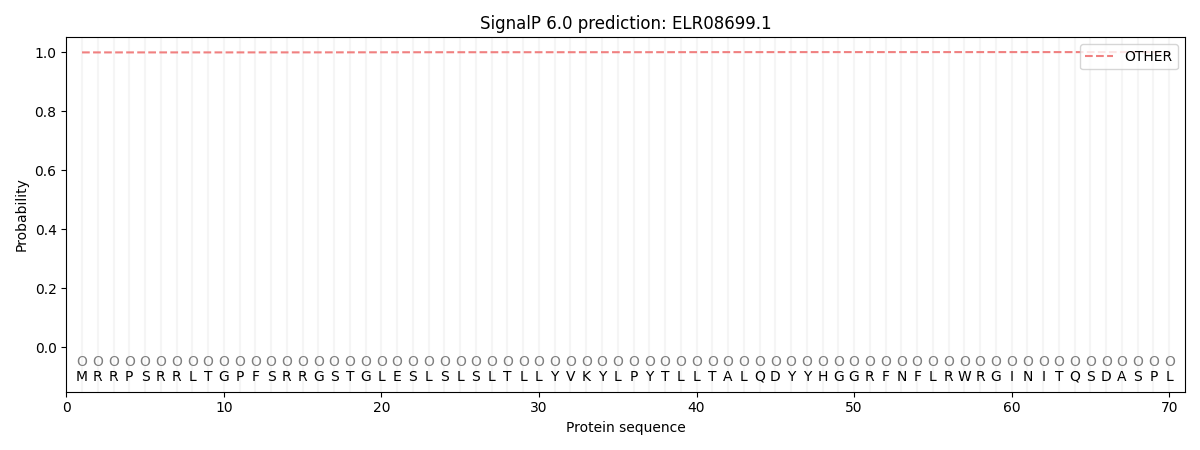
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999122 | 0.000895 |
