You are browsing environment: FUNGIDB
CAZyme Information: ELR07584.1
You are here: Home > Sequence: ELR07584.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Pseudogymnoascus destructans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Leotiomycetes; ; Pseudeurotiaceae; Pseudogymnoascus; Pseudogymnoascus destructans | |||||||||||
| CAZyme ID | ELR07584.1 | |||||||||||
| CAZy Family | GH72|CBM43 | |||||||||||
| CAZyme Description | Chitin-binding type-1 domain-containing protein [Source:UniProtKB/TrEMBL;Acc:L8G482] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA11 | 20 | 207 | 1.6e-64 | 0.9685863874345549 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 211316 | ChtBD1_1 | 4.25e-12 | 288 | 329 | 1 | 44 | Hevein or type 1 chitin binding domain; filamentous ascomycete subfamily. Hevein or type 1 chitin binding domain (ChtBD1), a lectin domain found in proteins from plants and fungi that bind N-acetylglucosamine, plant endochitinases, wound-induced proteins such as hevein, a major IgE-binding allergen in natural rubber latex, and the alpha subunit of Kluyveromyces lactis killer toxin. This domain is involved in the recognition and/or binding of chitin subunits; it typically occurs N-terminal to glycosyl hydrolase domains in chitinases, together with other carbohydrate-binding domains, or by itself in tandem-repeat arrangements. |
| 211311 | ChtBD1 | 4.63e-08 | 287 | 324 | 3 | 37 | Hevein or type 1 chitin binding domain. Hevein or type 1 chitin binding domain (ChtBD1), a lectin domain found in proteins from plants and fungi that bind N-acetylglucosamine, plant endochitinases, wound-induced proteins such as hevein, a major IgE-binding allergen in natural rubber latex, and the alpha subunit of Kluyveromyces lactis killer toxin. This domain is involved in the recognition and/or binding of chitin subunits; it typically occurs N-terminal to glycosyl hydrolase domains in chitinases, together with other carbohydrate-binding domains, or by itself in tandem-repeat arrangements. |
| 395135 | Chitin_bind_1 | 7.94e-08 | 288 | 321 | 1 | 34 | Chitin recognition protein. |
| 214593 | ChtBD1 | 4.15e-05 | 287 | 321 | 3 | 34 | Chitin binding domain. |
| 211312 | ChtBD1_GH19_hevein | 2.20e-04 | 288 | 321 | 2 | 35 | Hevein or Type 1 chitin binding domain subfamily co-occuring with family 19 glycosyl hydrolases or with barwin domains. This subfamily includes Hevein, a major IgE-binding allergen in natural rubber latex. ChtBD1 is a lectin domain found in proteins from plants and fungi that bind N-acetylglucosamine, plant endochitinases, wound-induced proteins, and the alpha subunit of Kluyveromyces lactis killer toxin. This domain is involved in the recognition and/or binding of chitin subunits; it typically occurs N-terminal to glycosyl hydrolase domains in chitinases, together with other carbohydrate-binding domains, or by itself in tandem-repeat arrangements. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 7.45e-82 | 19 | 221 | 19 | 224 | |
| 3.38e-81 | 1 | 257 | 1 | 260 | |
| 1.43e-80 | 19 | 218 | 18 | 220 | |
| 1.59e-80 | 1 | 257 | 1 | 260 | |
| 2.37e-80 | 19 | 218 | 18 | 220 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.03e-26 | 20 | 201 | 1 | 196 | Structure of Aspergillus oryzae AA11 Lytic Polysaccharide Monooxygenase with Zn [Aspergillus oryzae],4MAI_A Structure of Aspergillus oryzae AA11 Lytic Polysaccharide Monooxygenase with Cu(I) [Aspergillus oryzae] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
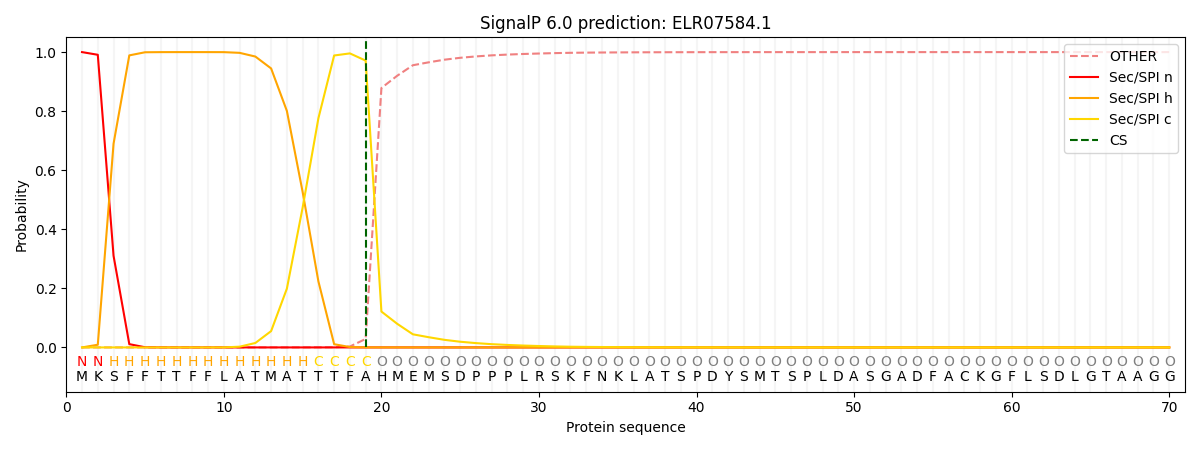
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000546 | 0.999436 | CS pos: 19-20. Pr: 0.9712 |
