You are browsing environment: FUNGIDB
CAZyme Information: ELR06038.1
You are here: Home > Sequence: ELR06038.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Pseudogymnoascus destructans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Leotiomycetes; ; Pseudeurotiaceae; Pseudogymnoascus; Pseudogymnoascus destructans | |||||||||||
| CAZyme ID | ELR06038.1 | |||||||||||
| CAZy Family | GH35 | |||||||||||
| CAZyme Description | Transmembrane 9 superfamily member [Source:UniProtKB/TrEMBL;Acc:L8FZY9] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL3 | 748 | 892 | 8.4e-52 | 0.7268041237113402 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397233 | EMP70 | 0.0 | 66 | 603 | 1 | 509 | Endomembrane protein 70. |
| 397360 | Pectate_lyase | 7.38e-68 | 746 | 886 | 12 | 146 | Pectate lyase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 6.12e-137 | 26 | 620 | 621 | 1209 | |
| 4.56e-85 | 641 | 885 | 12 | 254 | |
| 5.05e-83 | 633 | 885 | 3 | 245 | |
| 7.86e-50 | 750 | 886 | 33 | 162 | |
| 1.08e-49 | 750 | 886 | 33 | 162 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.35e-26 | 764 | 883 | 17 | 135 | Crystal Structure Of Pectate Lyase From Bacillus Sp. Strain Ksm-P15. [Bacillus sp. KSM-P15] |
|
| 2.69e-17 | 789 | 883 | 45 | 138 | The liganded structure of C. bescii family 3 pectate lyase [Caldicellulosiruptor bescii DSM 6725],4EW9_B The liganded structure of C. bescii family 3 pectate lyase [Caldicellulosiruptor bescii DSM 6725] |
|
| 2.75e-17 | 789 | 883 | 46 | 139 | The crystal structure of family 3 pectate lyase from Caldicellulosiruptor bescii [Caldicellulosiruptor bescii],3T9G_B The crystal structure of family 3 pectate lyase from Caldicellulosiruptor bescii [Caldicellulosiruptor bescii] |
|
| 2.06e-16 | 789 | 883 | 54 | 147 | C. bescii Family 3 pectate lyase double mutant K108A in complex with trigalacturonic acid [Caldicellulosiruptor bescii DSM 6725],4Z03_B C. bescii Family 3 pectate lyase double mutant K108A in complex with trigalacturonic acid [Caldicellulosiruptor bescii DSM 6725] |
|
| 2.06e-16 | 789 | 883 | 54 | 147 | C. bescii Family 3 pectate lyase double mutant K108A/E39Q in complex with trigalacturonic acid [Caldicellulosiruptor bescii DSM 6725],4YZ0_B C. bescii Family 3 pectate lyase double mutant K108A/E39Q in complex with trigalacturonic acid [Caldicellulosiruptor bescii DSM 6725] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7.65e-170 | 26 | 619 | 22 | 640 | Transmembrane 9 superfamily member 1 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=EMP70 PE=1 SV=2 |
|
| 9.65e-156 | 22 | 622 | 22 | 605 | Transmembrane 9 superfamily protein C1105.08 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=SPBC1105.08 PE=3 SV=1 |
|
| 1.45e-154 | 23 | 617 | 15 | 643 | Transmembrane 9 superfamily member 2 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=TMN2 PE=1 SV=1 |
|
| 2.35e-146 | 23 | 620 | 24 | 615 | Transmembrane 9 superfamily member 7 OS=Arabidopsis thaliana OX=3702 GN=TMN7 PE=2 SV=1 |
|
| 4.47e-146 | 21 | 620 | 30 | 636 | Transmembrane 9 superfamily member 2 OS=Mus musculus OX=10090 GN=Tm9sf2 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
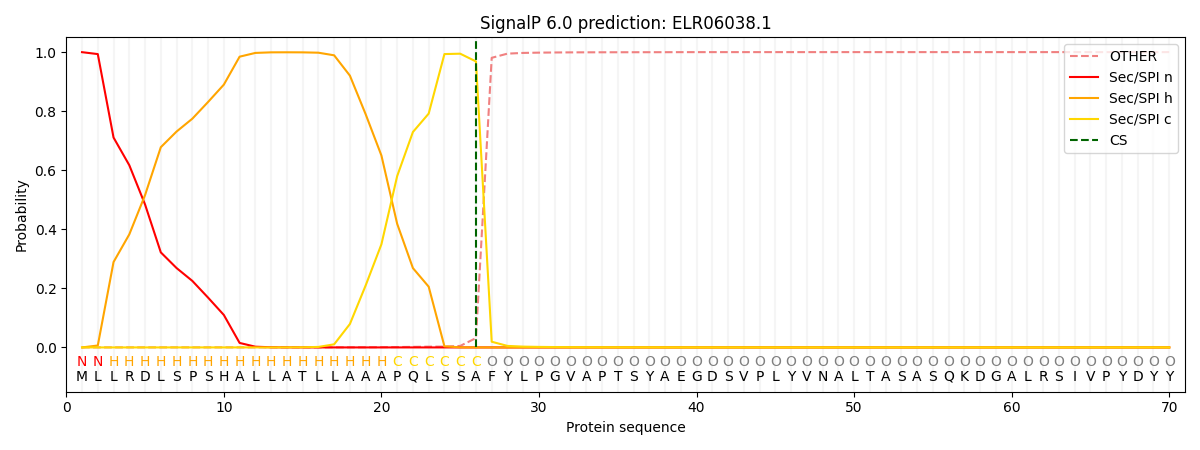
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000903 | 0.999052 | CS pos: 26-27. Pr: 0.9684 |
TMHMM Annotations download full data without filtering help
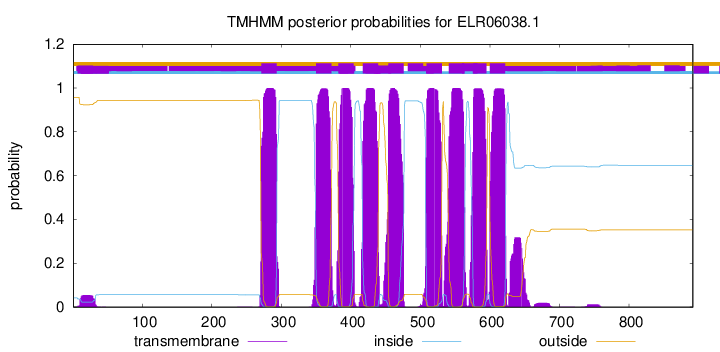
| Start | End |
|---|---|
| 271 | 293 |
| 350 | 372 |
| 382 | 404 |
| 417 | 439 |
| 454 | 476 |
| 509 | 531 |
| 541 | 563 |
| 576 | 595 |
| 600 | 622 |
