You are browsing environment: FUNGIDB
CAZyme Information: ELR03900.1
You are here: Home > Sequence: ELR03900.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Pseudogymnoascus destructans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Leotiomycetes; ; Pseudeurotiaceae; Pseudogymnoascus; Pseudogymnoascus destructans | |||||||||||
| CAZyme ID | ELR03900.1 | |||||||||||
| CAZy Family | GH13 | |||||||||||
| CAZyme Description | Mannosyltransferase [Source:UniProtKB/TrEMBL;Acc:L8FSE5] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.259:12 | 2.4.1.261:12 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT22 | 7 | 432 | 6e-89 | 0.9948586118251928 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 381301 | AKR_AKR7A1-5 | 4.51e-117 | 537 | 803 | 8 | 281 | AKR7A family of aldo-keto reductase (AKR). Aflatoxin B1 aldehyde reductase member 1/3 (AKR7A1/AKR7A3/AFAR) from Rattus norvegicus, aflatoxin B1 aldehyde reductase member 2 (AKR7A2/AFAR1/AFAR) and aflatoxin B1 aldehyde reductase member 3 (AKR7A3/AFAR2) from Homo sapiens, aflatoxin B1 aldehyde reductase member 2 (AKR7A2/AFAR2) from Rattus norvegicus, and aflatoxin B1 aldehyde reductase member 2 (AKR7A2/AKR7A5/AFAR) from Mus musculus, are founding members of aldo-keto reductase family 7 member A1-5 (AKR7A1-5), respectively. AKR7A2 (EC 1.1.1.n11), also called AFB1 aldehyde reductase 1, or AFB1-AR 1, or aldoketoreductase 7, or succinic semialdehyde reductase, or SSA reductase, catalyzes the NADPH-dependent reduction of succinic semialdehyde to gamma-hydroxybutyrate (GHB). It has NADPH-dependent aldehyde reductase activity towards 2-carboxybenzaldehyde, 2-nitrobenzaldehyde and pyridine-2-aldehyde (in vitro). AKR7A2, AKR7A3 (also called AFB1 aldehyde reductase 2 or AFB1-AR 2), and AKR7A4 (also called AFB1 aldehyde reductase 3, or AFB1-AR 3, or aldoketoreductase 7-like), may be involved in protection of liver against the toxic and carcinogenic effects of aflatoxin B1 (AFB1), a potent hepatocarcinogen. They can reduce the dialdehyde protein-binding form of AFB1 to the non-binding AFB1 dialcohol. |
| 281842 | Glyco_transf_22 | 1.81e-87 | 23 | 432 | 19 | 414 | Alg9-like mannosyltransferase family. Members of this family are mannosyltransferase enzymes. At least some members are localized in endoplasmic reticulum and involved in GPI anchor biosynthesis. |
| 395190 | Aldo_ket_red | 1.14e-38 | 537 | 805 | 6 | 270 | Aldo/keto reductase family. This family includes a number of K+ ion channel beta chain regulatory domains - these are reported to have oxidoreductase activity. |
| 223739 | Tas | 1.54e-37 | 530 | 804 | 7 | 287 | Predicted oxidoreductase (related to aryl-alcohol dehydrogenase) [General function prediction only]. |
| 381313 | AKR_AKR12A1_B1_C1 | 4.47e-36 | 530 | 799 | 7 | 283 | AKR12A, AKR12B, AKR12C families of aldo-keto reductase (AKR). Streptomyces fradiae TylCII, Saccharopolyspora erythraea EryBII, and Streptomyces avermitilis aveBVIII are founding members of aldo-keto reductase family 12 member A1 (AKR12A1), B1 (AKR12B1), and C1(AKR12C1), respectively. TylCII acts as a NDP-hexose 2,3-enoyl reductase. EryBII is a mycarose/desosamine reductase involved in L-mycarose and D-desosamine production. aveBVIII functions as a dTDP-4-keto-6-deoxy-L-hexose-2,3-reductase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.35e-250 | 2 | 538 | 39 | 575 | |
| 6.43e-250 | 2 | 538 | 35 | 570 | |
| 4.55e-249 | 2 | 538 | 35 | 570 | |
| 6.45e-249 | 2 | 538 | 35 | 570 | |
| 5.09e-221 | 2 | 532 | 29 | 566 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.06e-51 | 549 | 802 | 9 | 291 | Aflatoxin aldehyde reductase (AKR7A1) from Rat Liver [Rattus norvegicus],1GVE_B Aflatoxin aldehyde reductase (AKR7A1) from Rat Liver [Rattus norvegicus] |
|
| 6.83e-48 | 546 | 802 | 35 | 311 | Crystal structure of human aflatoxin B1 aldehyde reductase member 3 [Homo sapiens],2CLP_B Crystal structure of human aflatoxin B1 aldehyde reductase member 3 [Homo sapiens],2CLP_C Crystal structure of human aflatoxin B1 aldehyde reductase member 3 [Homo sapiens],2CLP_D Crystal structure of human aflatoxin B1 aldehyde reductase member 3 [Homo sapiens],2CLP_E Crystal structure of human aflatoxin B1 aldehyde reductase member 3 [Homo sapiens],2CLP_F Crystal structure of human aflatoxin B1 aldehyde reductase member 3 [Homo sapiens],2CLP_G Crystal structure of human aflatoxin B1 aldehyde reductase member 3 [Homo sapiens],2CLP_H Crystal structure of human aflatoxin B1 aldehyde reductase member 3 [Homo sapiens],2CLP_I Crystal structure of human aflatoxin B1 aldehyde reductase member 3 [Homo sapiens],2CLP_J Crystal structure of human aflatoxin B1 aldehyde reductase member 3 [Homo sapiens],2CLP_K Crystal structure of human aflatoxin B1 aldehyde reductase member 3 [Homo sapiens] |
|
| 1.60e-46 | 546 | 802 | 48 | 324 | Structure of the aflatoxin aldehyde reductase in complex with NADPH [Homo sapiens],2BP1_B Structure of the aflatoxin aldehyde reductase in complex with NADPH [Homo sapiens],2BP1_C Structure of the aflatoxin aldehyde reductase in complex with NADPH [Homo sapiens],2BP1_D Structure of the aflatoxin aldehyde reductase in complex with NADPH [Homo sapiens] |
|
| 7.50e-45 | 546 | 802 | 26 | 302 | mouse succinic semialdehyde reductase, AKR7A5 [Mus musculus],2C91_B mouse succinic semialdehyde reductase, AKR7A5 [Mus musculus],2C91_C mouse succinic semialdehyde reductase, AKR7A5 [Mus musculus],2C91_D mouse succinic semialdehyde reductase, AKR7A5 [Mus musculus],2C91_E mouse succinic semialdehyde reductase, AKR7A5 [Mus musculus],2C91_F mouse succinic semialdehyde reductase, AKR7A5 [Mus musculus],2C91_G mouse succinic semialdehyde reductase, AKR7A5 [Mus musculus],2C91_H mouse succinic semialdehyde reductase, AKR7A5 [Mus musculus],2C91_I mouse succinic semialdehyde reductase, AKR7A5 [Mus musculus],2C91_J mouse succinic semialdehyde reductase, AKR7A5 [Mus musculus] |
|
| 5.27e-22 | 553 | 777 | 34 | 266 | Structure of NADPH-dependent family 11 aldo-keto reductase AKR11A(apo) [Bacillus subtilis],1PZ0_A Structure of NADPH-dependent family 11 aldo-keto reductase AKR11A(holo) [Bacillus subtilis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.72e-110 | 4 | 539 | 12 | 542 | Alpha-1,2-mannosyltransferase alg9 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=alg9 PE=3 SV=1 |
|
| 3.16e-101 | 14 | 532 | 69 | 572 | Alpha-1,2-mannosyltransferase ALG9 OS=Homo sapiens OX=9606 GN=ALG9 PE=1 SV=2 |
|
| 4.90e-99 | 14 | 532 | 69 | 572 | Alpha-1,2-mannosyltransferase ALG9 OS=Mus musculus OX=10090 GN=Alg9 PE=2 SV=1 |
|
| 2.20e-76 | 558 | 804 | 1 | 262 | Aldo-keto reductase MYCFIDRAFT_156381 OS=Pseudocercospora fijiensis (strain CIRAD86) OX=383855 GN=MYCFIDRAFT_156381 PE=2 SV=1 |
|
| 1.32e-74 | 14 | 532 | 64 | 570 | Alpha-1,2-mannosyltransferase algn-9 OS=Caenorhabditis elegans OX=6239 GN=algn-9 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
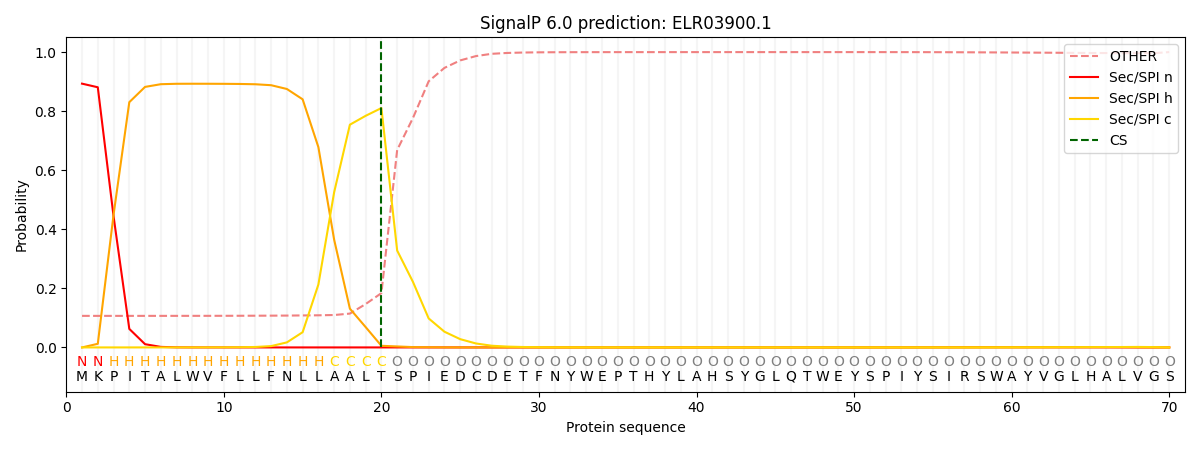
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.153599 | 0.846378 | CS pos: 20-21. Pr: 0.8105 |
TMHMM Annotations download full data without filtering help

| Start | End |
|---|---|
| 7 | 24 |
| 85 | 107 |
| 114 | 136 |
| 140 | 159 |
| 166 | 188 |
| 271 | 293 |
| 298 | 320 |
| 330 | 352 |
| 359 | 381 |
