You are browsing environment: FUNGIDB
CAZyme Information: ELR03522.1
Basic Information
help
| Species |
Pseudogymnoascus destructans
|
| Lineage |
Ascomycota; Leotiomycetes; ; Pseudeurotiaceae; Pseudogymnoascus; Pseudogymnoascus destructans
|
| CAZyme ID |
ELR03522.1
|
| CAZy Family |
CE4 |
| CAZyme Description |
unspecified product
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 534 |
|
58321.81 |
4.5315 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_Pdestructans20631-21 |
9225 |
658429 |
150 |
9075
|
|
| Gene Location |
No EC number prediction in ELR03522.1.
| Family |
Start |
End |
Evalue |
family coverage |
| GH76 |
66 |
455 |
1.4e-54 |
0.8212290502793296 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| 397638
|
Glyco_hydro_76 |
4.34e-13 |
92 |
444 |
43 |
340 |
Glycosyl hydrolase family 76. Family of alpha-1,6-mannanases. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 5.25e-152 |
48 |
527 |
71 |
564 |
| 4.35e-148 |
51 |
521 |
77 |
565 |
| 4.77e-146 |
40 |
530 |
67 |
572 |
| 3.34e-139 |
48 |
521 |
48 |
548 |
| 7.62e-139 |
24 |
533 |
71 |
661 |
ELR03522.1 has no PDB hit.
ELR03522.1 has no Swissprot hit.
This protein is predicted as SP
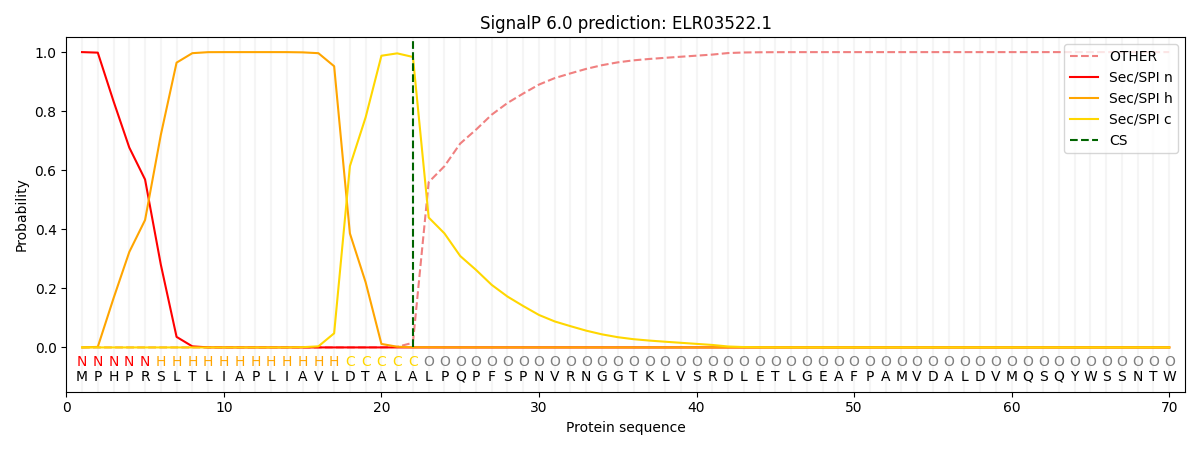
| Other |
SP_Sec_SPI |
CS Position |
| 0.000341 |
0.999620 |
CS pos: 22-23. Pr: 0.9835 |
There is no transmembrane helices in ELR03522.1.

