You are browsing environment: FUNGIDB
CAZyme Information: EKG22208.1
You are here: Home > Sequence: EKG22208.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Macrophomina phaseolina | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Botryosphaeriaceae; Macrophomina; Macrophomina phaseolina | |||||||||||
| CAZyme ID | EKG22208.1 | |||||||||||
| CAZy Family | GT8 | |||||||||||
| CAZyme Description | Chitinase [Source:UniProtKB/TrEMBL;Acc:K2S5L2] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH18 | 49 | 383 | 8.6e-69 | 0.9662162162162162 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 282020 | Vps53_N | 5.48e-108 | 386 | 764 | 2 | 372 | Vps53-like, N-terminal. Vps53 complexes with Vps52 and Vps54 to form a multi- subunit complex involved in regulating membrane trafficking events. |
| 119365 | GH18_chitinase | 2.43e-97 | 48 | 371 | 1 | 322 | The GH18 (glycosyl hydrolases, family 18) type II chitinases hydrolyze chitin, an abundant polymer of N-acetylglucosamine and have been identified in bacteria, fungi, insects, plants, viruses, and protozoan parasites. The structure of this domain is an eight-stranded alpha/beta barrel with a pronounced active-site cleft at the C-terminal end of the beta-barrel. |
| 214753 | Glyco_18 | 3.02e-83 | 49 | 371 | 3 | 334 | Glyco_18 domain. |
| 395573 | Glyco_hydro_18 | 2.28e-75 | 49 | 371 | 3 | 307 | Glycosyl hydrolases family 18. |
| 225862 | ChiA | 2.98e-66 | 46 | 372 | 38 | 424 | Chitinase, GH18 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 38 | 1226 | 15 | 1221 | |
| 0.0 | 45 | 1228 | 20 | 1155 | |
| 1.56e-161 | 38 | 386 | 15 | 372 | |
| 6.32e-156 | 36 | 385 | 14 | 390 | |
| 1.83e-155 | 38 | 385 | 16 | 372 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.43e-60 | 50 | 377 | 7 | 354 | Chain A, CHITINASE 1 [Coccidioides immitis],1LL4_A Chain A, Chitinase 1 [Coccidioides immitis],1LL4_B Chain B, Chitinase 1 [Coccidioides immitis],1LL4_C Chain C, Chitinase 1 [Coccidioides immitis],1LL4_D Chain D, Chitinase 1 [Coccidioides immitis] |
|
| 1.62e-59 | 50 | 377 | 7 | 354 | Chain A, CHITINASE 1 [Coccidioides immitis],1LL7_B Chain B, CHITINASE 1 [Coccidioides immitis] |
|
| 2.99e-59 | 50 | 377 | 7 | 354 | Chain A, CHITINASE 1 [Coccidioides immitis],1LL6_B Chain B, CHITINASE 1 [Coccidioides immitis],1LL6_C Chain C, CHITINASE 1 [Coccidioides immitis],1LL6_D Chain D, CHITINASE 1 [Coccidioides immitis] |
|
| 1.57e-54 | 51 | 377 | 21 | 367 | Crystal structure of Aspergillus niger chitinase B [Aspergillus niger] |
|
| 5.71e-53 | 50 | 377 | 9 | 357 | Crystal structure of a native chitinase from Aspergillus fumigatus YJ-407 [Aspergillus fumigatus],1WNO_B Crystal structure of a native chitinase from Aspergillus fumigatus YJ-407 [Aspergillus fumigatus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.43e-117 | 386 | 1081 | 39 | 796 | Vacuolar protein sorting-associated protein 53 homolog OS=Gallus gallus OX=9031 GN=VPS53 PE=2 SV=1 |
|
| 9.02e-114 | 386 | 1081 | 40 | 797 | Vacuolar protein sorting-associated protein 53 homolog OS=Mus musculus OX=10090 GN=Vps53 PE=1 SV=1 |
|
| 6.39e-113 | 386 | 1081 | 40 | 797 | Vacuolar protein sorting-associated protein 53 homolog OS=Homo sapiens OX=9606 GN=VPS53 PE=1 SV=2 |
|
| 6.39e-113 | 386 | 1081 | 40 | 797 | Vacuolar protein sorting-associated protein 53 homolog OS=Pongo abelii OX=9601 GN=VPS53 PE=2 SV=2 |
|
| 8.66e-99 | 485 | 1074 | 116 | 695 | Vacuolar protein sorting-associated protein 53 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=vps53 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
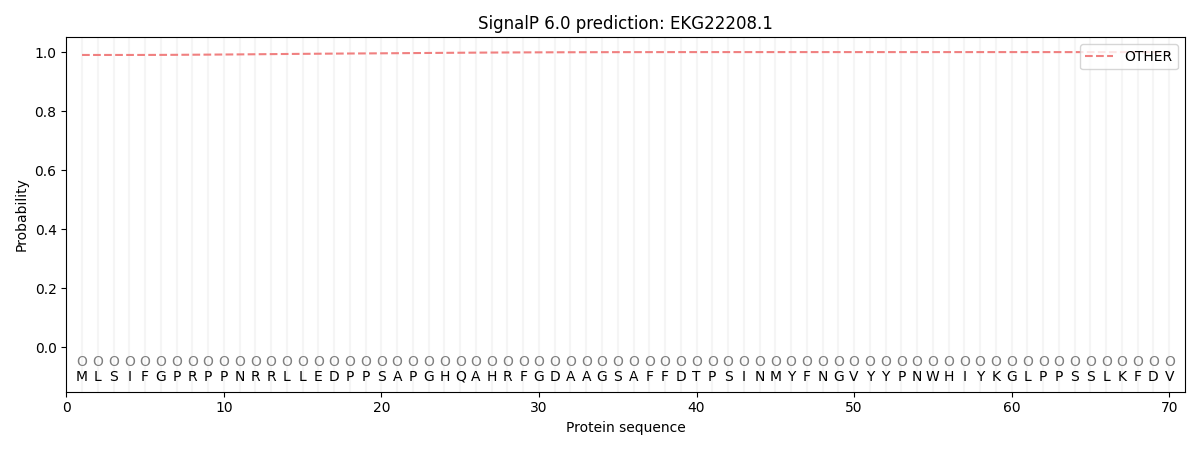
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.991107 | 0.008917 |
