You are browsing environment: FUNGIDB
CAZyme Information: EKG20603.1
You are here: Home > Sequence: EKG20603.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Macrophomina phaseolina | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Botryosphaeriaceae; Macrophomina; Macrophomina phaseolina | |||||||||||
| CAZyme ID | EKG20603.1 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | Cutinase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE5 | 44 | 228 | 6.4e-40 | 0.9894179894179894 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395860 | Cutinase | 4.63e-45 | 44 | 229 | 1 | 173 | Cutinase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.69e-50 | 10 | 229 | 5 | 208 | |
| 1.96e-48 | 43 | 231 | 224 | 397 | |
| 1.98e-45 | 34 | 229 | 176 | 355 | |
| 9.35e-45 | 24 | 230 | 82 | 275 | |
| 1.84e-44 | 34 | 229 | 184 | 363 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.90e-25 | 45 | 225 | 79 | 243 | Structure of cutinase from Trichoderma reesei in its native form. [Trichoderma reesei QM6a],4PSD_A Structure of Trichoderma reesei cutinase native form. [Trichoderma reesei QM6a],4PSE_A Trichoderma reesei cutinase in complex with a C11Y4 phosphonate inhibitor [Trichoderma reesei QM6a],4PSE_B Trichoderma reesei cutinase in complex with a C11Y4 phosphonate inhibitor [Trichoderma reesei QM6a] |
|
| 2.76e-19 | 44 | 186 | 30 | 178 | Chain A, CUTINASE [Fusarium vanettenii] |
|
| 4.05e-19 | 44 | 186 | 16 | 164 | The 1.15 angstrom refined structure of fusarium solani pisi cutinase [Fusarium vanettenii] |
|
| 5.37e-19 | 44 | 186 | 30 | 178 | Chain A, CUTINASE [Fusarium vanettenii] |
|
| 5.37e-19 | 44 | 186 | 30 | 178 | Chain A, CUTINASE [Fusarium vanettenii] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.10e-44 | 27 | 230 | 41 | 222 | Cutinase 4 OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=cut4 PE=2 SV=1 |
|
| 1.03e-33 | 28 | 230 | 14 | 201 | Cutinase OS=Botryotinia fuckeliana OX=40559 GN=cutA PE=1 SV=1 |
|
| 3.97e-33 | 44 | 231 | 30 | 201 | Cutinase OS=Monilinia fructicola OX=38448 GN=CUT1 PE=2 SV=1 |
|
| 6.35e-30 | 31 | 229 | 53 | 236 | Cutinase OS=Blumeria hordei OX=2867405 GN=CUT1 PE=3 SV=1 |
|
| 1.70e-28 | 5 | 229 | 5 | 215 | Probable cutinase 3 OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=AFUB_099910 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
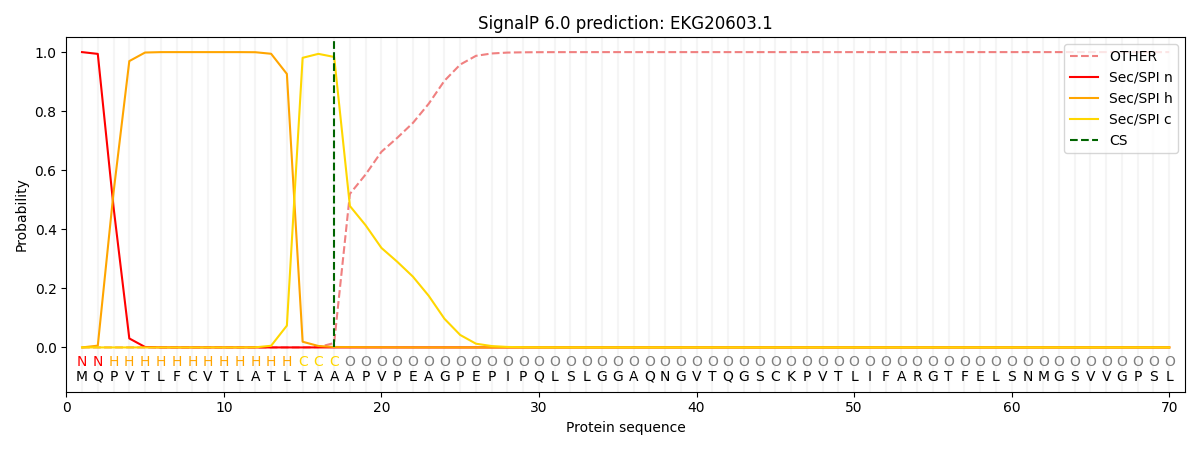
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000219 | 0.999750 | CS pos: 17-18. Pr: 0.9836 |
