You are browsing environment: FUNGIDB
CAZyme Information: EKG17011.1
You are here: Home > Sequence: EKG17011.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Macrophomina phaseolina | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Botryosphaeriaceae; Macrophomina; Macrophomina phaseolina | |||||||||||
| CAZyme ID | EKG17011.1 | |||||||||||
| CAZy Family | GH35 | |||||||||||
| CAZyme Description | Chitin-binding type 1 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA7 | 135 | 345 | 1.2e-76 | 0.4519650655021834 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396238 | FAD_binding_4 | 2.50e-26 | 143 | 277 | 1 | 136 | FAD binding domain. This family consists of various enzymes that use FAD as a co-factor, most of the enzymes are similar to oxygen oxidoreductase. One of the enzymes Vanillyl-alcohol oxidase (VAO) has a solved structure, the alignment includes the FAD binding site, called the PP-loop, between residues 99-110. The FAD molecule is covalently bound in the known structure, however the residue that links to the FAD is not in the alignment. VAO catalyzes the oxidation of a wide variety of substrates, ranging form aromatic amines to 4-alkylphenols. Other members of this family include D-lactate dehydrogenase, this enzyme catalyzes the conversion of D-lactate to pyruvate using FAD as a co-factor; mitomycin radical oxidase, this enzyme oxidizes the reduced form of mitomycins and is involved in mitomycin resistance. This family includes MurB an UDP-N-acetylenolpyruvoylglucosamine reductase enzyme EC:1.1.1.158. This enzyme is involved in the biosynthesis of peptidoglycan. |
| 223354 | GlcD | 1.67e-23 | 140 | 369 | 29 | 267 | FAD/FMN-containing dehydrogenase [Energy production and conversion]. |
| 211316 | ChtBD1_1 | 9.60e-16 | 36 | 74 | 1 | 39 | Hevein or type 1 chitin binding domain; filamentous ascomycete subfamily. Hevein or type 1 chitin binding domain (ChtBD1), a lectin domain found in proteins from plants and fungi that bind N-acetylglucosamine, plant endochitinases, wound-induced proteins such as hevein, a major IgE-binding allergen in natural rubber latex, and the alpha subunit of Kluyveromyces lactis killer toxin. This domain is involved in the recognition and/or binding of chitin subunits; it typically occurs N-terminal to glycosyl hydrolase domains in chitinases, together with other carbohydrate-binding domains, or by itself in tandem-repeat arrangements. |
| 211311 | ChtBD1 | 1.59e-10 | 36 | 74 | 1 | 37 | Hevein or type 1 chitin binding domain. Hevein or type 1 chitin binding domain (ChtBD1), a lectin domain found in proteins from plants and fungi that bind N-acetylglucosamine, plant endochitinases, wound-induced proteins such as hevein, a major IgE-binding allergen in natural rubber latex, and the alpha subunit of Kluyveromyces lactis killer toxin. This domain is involved in the recognition and/or binding of chitin subunits; it typically occurs N-terminal to glycosyl hydrolase domains in chitinases, together with other carbohydrate-binding domains, or by itself in tandem-repeat arrangements. |
| 395135 | Chitin_bind_1 | 1.88e-10 | 36 | 73 | 1 | 36 | Chitin recognition protein. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.50e-169 | 13 | 534 | 15 | 594 | |
| 4.53e-166 | 28 | 525 | 147 | 660 | |
| 5.12e-165 | 28 | 525 | 147 | 660 | |
| 4.62e-164 | 31 | 534 | 30 | 516 | |
| 2.02e-135 | 109 | 526 | 43 | 463 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.34e-78 | 109 | 529 | 27 | 436 | Chain AAA, Chitooligosaccharide oxidase [Fusarium graminearum PH-1] |
|
| 7.89e-77 | 109 | 493 | 27 | 403 | Xylooligosaccharide oxidase from Myceliophthora thermophila C1 [Thermothelomyces thermophilus ATCC 42464],5L6F_A Xylooligosaccharide oxidase from Myceliophthora thermophila C1 in complex with Xylobiose [Thermothelomyces thermophilus ATCC 42464],5L6G_A Xylooligosaccharide oxidase from Myceliophthora thermophila C1 in complex with Xylose [Thermothelomyces thermophilus ATCC 42464] |
|
| 2.01e-73 | 112 | 534 | 6 | 422 | Crystal structure of carbohydrate oxidase from Microdochium nivale [Microdochium nivale],3RJA_A Crystal structure of carbohydrate oxidase from Microdochium nivale in complex with substrate analogue [Microdochium nivale] |
|
| 2.73e-73 | 109 | 529 | 35 | 449 | Chain A, FAD-binding PCMH-type domain-containing protein [Fusarium graminearum PH-1] |
|
| 3.99e-68 | 112 | 530 | 12 | 425 | The crystal structure of an Acremonium strictum glucooligosaccharide oxidase reveals a novel flavinylation [Sarocladium strictum],2AXR_A Crystal structure of glucooligosaccharide oxidase from Acremonium strictum: a novel flavinylation of 6-S-cysteinyl, 8alpha-N1-histidyl FAD [Sarocladium strictum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.20e-77 | 109 | 529 | 27 | 436 | Chitooligosaccharide oxidase OS=Gibberella zeae (strain ATCC MYA-4620 / CBS 123657 / FGSC 9075 / NRRL 31084 / PH-1) OX=229533 GN=chitO PE=1 SV=1 |
|
| 4.06e-76 | 109 | 493 | 27 | 403 | Xylooligosaccharide oxidase OS=Myceliophthora thermophila (strain ATCC 42464 / BCRC 31852 / DSM 1799) OX=573729 GN=xylO PE=1 SV=1 |
|
| 1.81e-72 | 112 | 534 | 28 | 444 | Carbohydrate oxidase OS=Microdochium nivale OX=5520 GN=MnCO PE=1 SV=2 |
|
| 1.86e-67 | 112 | 530 | 31 | 444 | Glucooligosaccharide oxidase OS=Sarocladium strictum OX=5046 GN=gluO PE=1 SV=1 |
|
| 7.02e-65 | 112 | 531 | 32 | 443 | FAD-linked oxidoreductase dpmaF OS=Metarhizium anisopliae OX=5530 GN=dpmaF PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
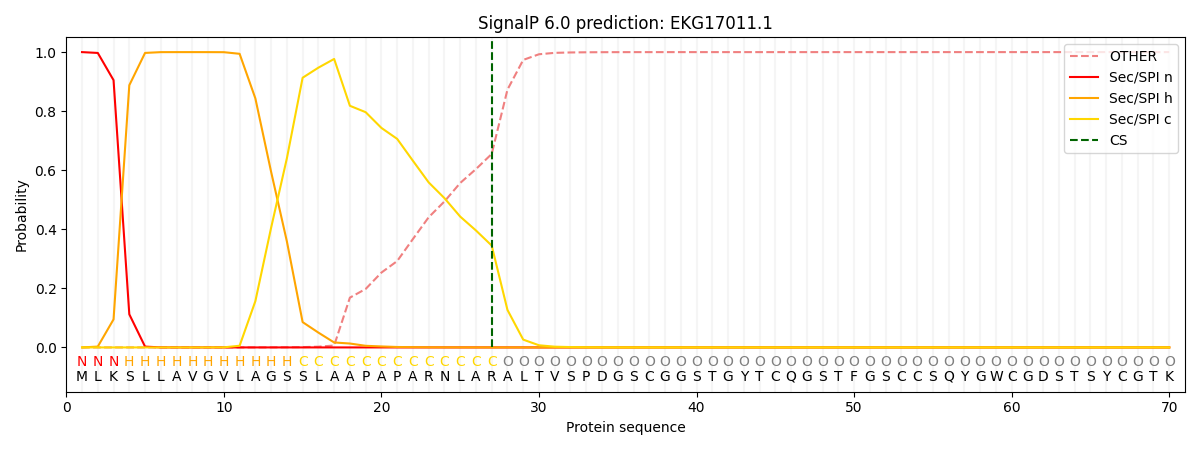
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000327 | 0.999643 | CS pos: 27-28. Pr: 0.3448 |
