You are browsing environment: FUNGIDB
CAZyme Information: EKG16430.1
You are here: Home > Sequence: EKG16430.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Macrophomina phaseolina | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Botryosphaeriaceae; Macrophomina; Macrophomina phaseolina | |||||||||||
| CAZyme ID | EKG16430.1 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | Six-hairpin glycosidase-like protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH146 | 31 | 549 | 5.3e-192 | 0.9960238568588469 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 400342 | Glyco_hydro_127 | 2.50e-164 | 31 | 550 | 1 | 503 | Beta-L-arabinofuranosidase, GH127. One member of this family, from Bidobacterium longicum, UniProtKB:E8MGH8, has been characterized as an unusual beta-L-arabinofuranosidase enzyme, EC:3.2.1.185. It rleases l-arabinose from the l-arabinofuranose (Araf)-beta1,2-Araf disaccharide and also transglycosylates 1-alkanols with retention of the anomeric configuration. Terminal beta-l-arabinofuranosyl residues have been found in arabinogalactan proteins from a mumber of different plantt species. beta-l-Arabinofuranosyl linkages with 1-4 arabinofuranosides are also found in the sugar chains of extensin and solanaceous lectins, hydroxyproline (Hyp)2-rich glycoproteins that are widely observed in plant cell wall fractions. The critical residue for catalytic activity is Glu-338, in a ET/SCAS sequence context. |
| 226063 | COG3533 | 1.33e-105 | 32 | 554 | 12 | 505 | Uncharacterized conserved protein, DUF1680 family [Function unknown]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.25e-311 | 4 | 607 | 17 | 621 | |
| 3.49e-310 | 10 | 609 | 32 | 623 | |
| 5.72e-310 | 4 | 609 | 26 | 631 | |
| 5.83e-307 | 7 | 609 | 33 | 634 | |
| 6.38e-306 | 5 | 609 | 31 | 634 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.80e-89 | 27 | 555 | 28 | 565 | Chain AAA, Acetyl-CoA carboxylase, biotin carboxylase [Bacteroides thetaiotaomicron VPI-5482] |
|
| 1.15e-84 | 27 | 555 | 28 | 565 | Beta-L-arabinofuranosidase [Bacteroides thetaiotaomicron] |
|
| 1.44e-12 | 157 | 550 | 183 | 560 | Crystal structure of native Ara127N, a GH127 beta-L-arabinofuranosidase from Geobacillus Stearothermophilus T6 [Geobacillus stearothermophilus],4QJY_B Crystal structure of native Ara127N, a GH127 beta-L-arabinofuranosidase from Geobacillus Stearothermophilus T6 [Geobacillus stearothermophilus] |
|
| 7.99e-12 | 161 | 550 | 192 | 607 | Glycoside hydrolase BT_1003 [Bacteroides thetaiotaomicron] |
|
| 2.30e-11 | 157 | 550 | 183 | 560 | Crystal structure of Ara127N-Se, a GH127 beta-L-arabinofuranosidase from Geobacillus Stearothermophilus T6 [Geobacillus stearothermophilus],4QK0_B Crystal structure of Ara127N-Se, a GH127 beta-L-arabinofuranosidase from Geobacillus Stearothermophilus T6 [Geobacillus stearothermophilus],4QK0_C Crystal structure of Ara127N-Se, a GH127 beta-L-arabinofuranosidase from Geobacillus Stearothermophilus T6 [Geobacillus stearothermophilus],4QK0_D Crystal structure of Ara127N-Se, a GH127 beta-L-arabinofuranosidase from Geobacillus Stearothermophilus T6 [Geobacillus stearothermophilus] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as OTHER
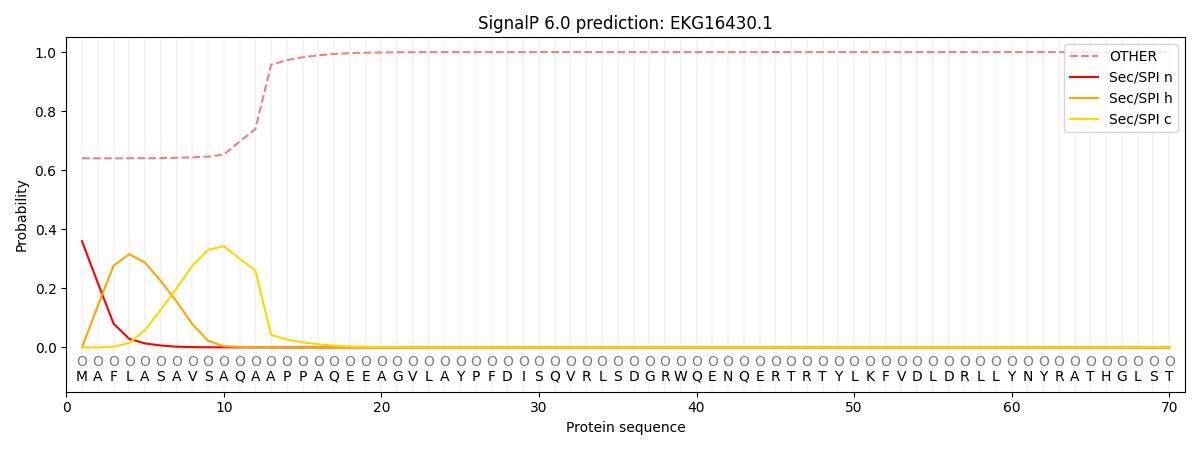
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.648408 | 0.351578 |
