You are browsing environment: FUNGIDB
EKG12373.1
Basic Information
help
Species
Macrophomina phaseolina
Lineage
Ascomycota; Dothideomycetes; ; Botryosphaeriaceae; Macrophomina; Macrophomina phaseolina
CAZyme ID
EKG12373.1
CAZy Family
AA9
CAZyme Description
Lactonase 7-bladed beta propeller
CAZyme Property
Protein Length
CGC
Molecular Weight
Isoelectric Point
337
AHHD01000451|CGC4 38932.86
6.7386
Genome Property
Genome Version/Assembly ID
Genes
Strain NCBI Taxon ID
Non Protein Coding Genes
Protein Coding Genes
FungiDB-61_MphaseolinaMS6
14712
1126212
906
13806
Gene Location
No EC number prediction in EKG12373.1.
Family
Start
End
Evalue
family coverage
GT34
185
286
7.6e-19
0.43089430894308944
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
368535
Glyco_transf_34
7.57e-13
117
285
38
231
galactosyl transferase GMA12/MNN10 family. This family contains a number of glycosyltransferase enzymes that contain a DXD motif. This family includes a number of C. elegans homologs where the DXD is replaced by DXH. Some members of this family are included in glycosyltransferase family 34.
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
1.46e-86
87
324
143
383
1.46e-86
87
324
143
383
1.46e-86
87
324
143
383
1.46e-86
87
324
143
383
2.07e-83
87
326
175
417
EKG12373.1 has no PDB hit.
EKG12373.1 has no Swissprot hit.
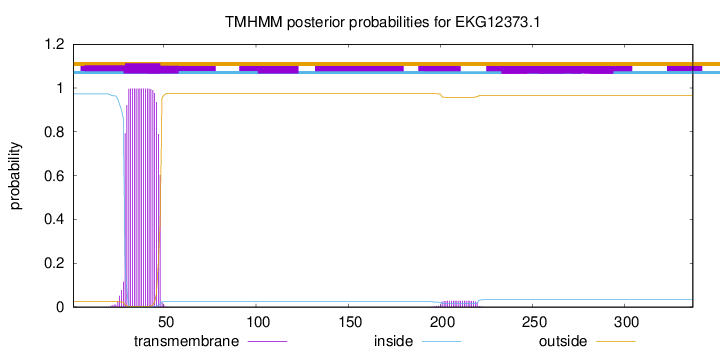
This protein is predicted as OTHER
Other
SP_Sec_SPI
CS Position
0.999702
0.000323