You are browsing environment: FUNGIDB
CAZyme Information: EJT79928.1
You are here: Home > Sequence: EJT79928.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
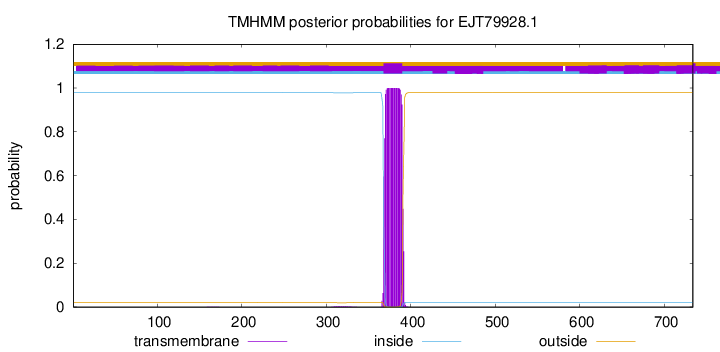
TMHMM annotations
Basic Information help
| Species | Gaeumannomyces tritici | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Magnaporthaceae; Gaeumannomyces; Gaeumannomyces tritici | |||||||||||
| CAZyme ID | EJT79928.1 | |||||||||||
| CAZy Family | GH94 | |||||||||||
| CAZyme Description | endo-beta-1,3-glucanase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH17 | 454 | 720 | 5.4e-26 | 0.9485530546623794 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 227625 | Scw11 | 1.66e-41 | 456 | 717 | 60 | 305 | Exo-beta-1,3-glucanase, GH17 family [Carbohydrate transport and metabolism]. |
| 366033 | Glyco_hydro_17 | 2.24e-04 | 459 | 720 | 25 | 304 | Glycosyl hydrolases family 17. |
| 185594 | PTZ00395 | 0.008 | 54 | 278 | 298 | 484 | Sec24-related protein; Provisional |
| 236090 | PRK07764 | 0.009 | 9 | 186 | 633 | 807 | DNA polymerase III subunits gamma and tau; Validated |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.20e-270 | 61 | 734 | 83 | 767 | |
| 7.56e-201 | 1 | 732 | 1 | 729 | |
| 1.49e-197 | 4 | 731 | 7 | 715 | |
| 1.49e-197 | 4 | 731 | 7 | 715 | |
| 2.98e-197 | 4 | 731 | 7 | 715 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.40e-25 | 456 | 725 | 50 | 294 | Crystal structure of glycoside hydrolase family 17 beta-1,3-glucanosyltransferase from Rhizomucor miehei [Rhizomucor miehei CAU432] |
|
| 8.60e-25 | 456 | 725 | 50 | 294 | Active-site mutant of Rhizomucor miehei beta-1,3-glucanosyltransferase in complex with laminaribiose [Rhizomucor miehei CAU432],4WTS_A Active-site mutant of Rhizomucor miehei beta-1,3-glucanosyltransferase in complex with laminaritriose [Rhizomucor miehei CAU432] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 9.60e-149 | 108 | 733 | 32 | 687 | Probable glucan endo-1,3-beta-glucosidase btgC OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=btgC PE=3 SV=1 |
|
| 1.69e-148 | 102 | 733 | 35 | 684 | Probable glucan endo-1,3-beta-glucosidase btgC OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=btgC PE=3 SV=2 |
|
| 1.69e-148 | 102 | 733 | 35 | 684 | Probable glucan endo-1,3-beta-glucosidase btgC OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=btgC PE=3 SV=1 |
|
| 1.15e-146 | 108 | 733 | 32 | 687 | Probable glucan endo-1,3-beta-glucosidase btgC OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=btgC PE=3 SV=1 |
|
| 2.28e-146 | 108 | 733 | 32 | 687 | Probable glucan endo-1,3-beta-glucosidase btgC OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=btgC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
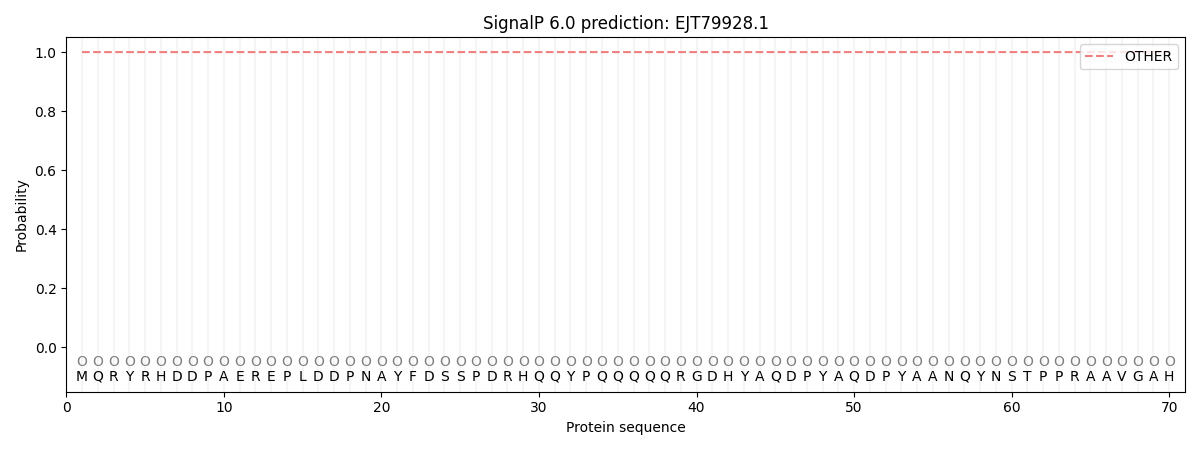
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000023 | 0.000005 |