You are browsing environment: FUNGIDB
CAZyme Information: EJT75856.1
You are here: Home > Sequence: EJT75856.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
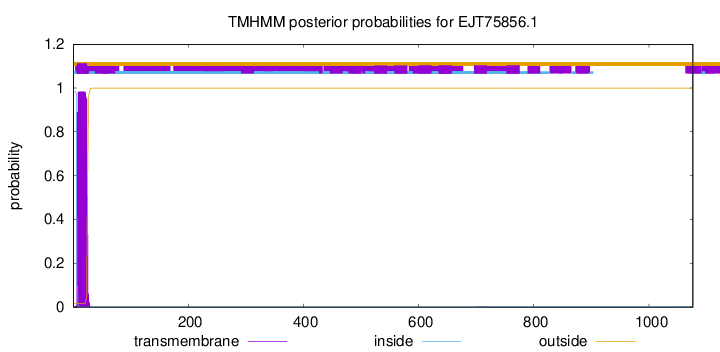
TMHMM annotations
Basic Information help
| Species | Gaeumannomyces tritici | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Magnaporthaceae; Gaeumannomyces; Gaeumannomyces tritici | |||||||||||
| CAZyme ID | EJT75856.1 | |||||||||||
| CAZy Family | GH20 | |||||||||||
| CAZyme Description | alpha-1,2-Mannosidase [Source:UniProtKB/TrEMBL;Acc:J3NWX4] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.113:7 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH47 | 268 | 1074 | 5e-172 | 0.9955156950672646 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396217 | Glyco_hydro_47 | 1.64e-167 | 267 | 1074 | 1 | 453 | Glycosyl hydrolase family 47. Members of this family are alpha-mannosidases that catalyze the hydrolysis of the terminal 1,2-linked alpha-D-mannose residues in the oligo-mannose oligosaccharide Man(9)(GlcNAc)(2). |
| 240427 | PTZ00470 | 3.55e-74 | 211 | 1075 | 33 | 519 | glycoside hydrolase family 47 protein; Provisional |
| 223021 | PHA03247 | 5.14e-05 | 59 | 260 | 2739 | 2938 | large tegument protein UL36; Provisional |
| 223065 | PHA03378 | 6.77e-05 | 39 | 242 | 597 | 801 | EBNA-3B; Provisional |
| 411408 | PspC_subgroup_2 | 7.12e-05 | 51 | 240 | 284 | 455 | pneumococcal surface protein PspC, LPXTG-anchored form. The pneumococcal surface protein PspC, as described in Streptococcus pneumoniae, is a repetitive and highly variable protein, recognized by a conserved N-terminal domain and also by genomic location. This form, subgroup 2, is anchored covalently after cleavage by sortase at a C-terminal LPXTG site. The other form, subgroup 1, has variable numbers of a choline-binding repeat in the C-terminal region, and is also known as choline-binding protein A. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 1075 | 1 | 1024 | |
| 5.01e-301 | 2 | 1076 | 4 | 1043 | |
| 1.94e-282 | 5 | 1075 | 5 | 1026 | |
| 1.13e-267 | 90 | 1076 | 45 | 882 | |
| 1.28e-259 | 207 | 1076 | 142 | 898 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.10e-32 | 271 | 445 | 16 | 181 | Crystal structure of the class I human endoplasmic reticulum 1,2-alpha-mannosidase and Man9GlcNAc2-PA complex [Homo sapiens] |
|
| 3.24e-32 | 271 | 445 | 16 | 181 | Crystal structure of the class I human endoplasmic reticulum 1,2-alpha-mannosidase T688A mutant and Thio-disaccharide substrate analog complex [Homo sapiens],5KK7_B Crystal structure of the class I human endoplasmic reticulum 1,2-alpha-mannosidase T688A mutant and Thio-disaccharide substrate analog complex [Homo sapiens] |
|
| 3.39e-32 | 271 | 445 | 21 | 186 | Crystal Structure Of Human Class I Alpha1,2-Mannosidase [Homo sapiens] |
|
| 3.50e-32 | 271 | 445 | 21 | 186 | Crystal Structure Of Human Class I Alpha1,2-Mannosidase In Complex With 1-Deoxymannojirimycin [Homo sapiens],1FO3_A Crystal Structure Of Human Class I Alpha1,2-Mannosidase In Complex With Kifunensine [Homo sapiens] |
|
| 9.67e-32 | 271 | 445 | 99 | 264 | Crystal Structure Of Human Class I alpha-1,2-Mannosidase In Complex With Thio-Disaccharide Substrate Analogue [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.30e-32 | 161 | 1071 | 70 | 623 | Mannosyl-oligosaccharide 1,2-alpha-mannosidase IB OS=Mus musculus OX=10090 GN=Man1a2 PE=1 SV=1 |
|
| 7.10e-31 | 254 | 1071 | 178 | 623 | Mannosyl-oligosaccharide 1,2-alpha-mannosidase IB OS=Homo sapiens OX=9606 GN=MAN1A2 PE=1 SV=1 |
|
| 1.79e-30 | 271 | 445 | 260 | 425 | Endoplasmic reticulum mannosyl-oligosaccharide 1,2-alpha-mannosidase OS=Homo sapiens OX=9606 GN=MAN1B1 PE=1 SV=2 |
|
| 3.00e-30 | 260 | 479 | 98 | 306 | Mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS2 OS=Arabidopsis thaliana OX=3702 GN=MNS2 PE=1 SV=1 |
|
| 5.78e-30 | 271 | 481 | 218 | 416 | Endoplasmic reticulum mannosyl-oligosaccharide 1,2-alpha-mannosidase OS=Rattus norvegicus OX=10116 GN=Man1b1 PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
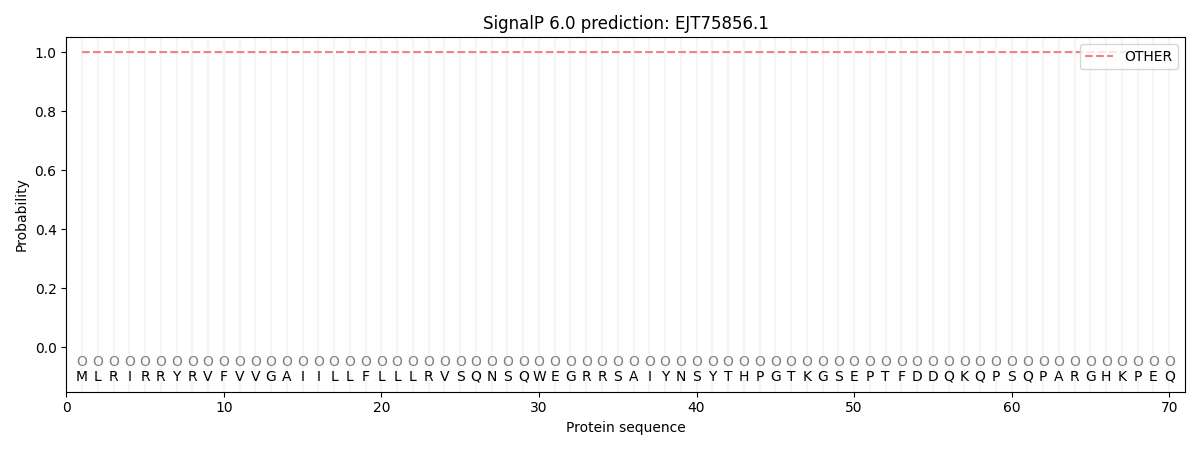
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000019 | 0.000008 |