You are browsing environment: FUNGIDB
CAZyme Information: EJT74865.1
You are here: Home > Sequence: EJT74865.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Gaeumannomyces tritici | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Magnaporthaceae; Gaeumannomyces; Gaeumannomyces tritici | |||||||||||
| CAZyme ID | EJT74865.1 | |||||||||||
| CAZy Family | GH16 | |||||||||||
| CAZyme Description | FAD-binding PCMH-type domain-containing protein [Source:UniProtKB/TrEMBL;Acc:J3P5B4] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA7 | 127 | 309 | 3.9e-48 | 0.3777292576419214 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 223354 | GlcD | 1.06e-20 | 85 | 549 | 7 | 450 | FAD/FMN-containing dehydrogenase [Energy production and conversion]. |
| 396238 | FAD_binding_4 | 3.01e-20 | 125 | 265 | 5 | 139 | FAD binding domain. This family consists of various enzymes that use FAD as a co-factor, most of the enzymes are similar to oxygen oxidoreductase. One of the enzymes Vanillyl-alcohol oxidase (VAO) has a solved structure, the alignment includes the FAD binding site, called the PP-loop, between residues 99-110. The FAD molecule is covalently bound in the known structure, however the residue that links to the FAD is not in the alignment. VAO catalyzes the oxidation of a wide variety of substrates, ranging form aromatic amines to 4-alkylphenols. Other members of this family include D-lactate dehydrogenase, this enzyme catalyzes the conversion of D-lactate to pyruvate using FAD as a co-factor; mitomycin radical oxidase, this enzyme oxidizes the reduced form of mitomycins and is involved in mitomycin resistance. This family includes MurB an UDP-N-acetylenolpyruvoylglucosamine reductase enzyme EC:1.1.1.158. This enzyme is involved in the biosynthesis of peptidoglycan. |
| 369658 | BBE | 5.46e-13 | 512 | 550 | 1 | 39 | Berberine and berberine like. This domain is found in the berberine bridge and berberine bridge- like enzymes which are involved in the biosynthesis of numerous isoquinoline alkaloids. They catalyze the transformation of the N-methyl group of (S)-reticuline into the C-8 berberine bridge carbon of (S)-scoulerine. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 4.16e-66 | 29 | 570 | 500 | 1065 | |
| 4.16e-66 | 29 | 570 | 500 | 1065 | |
| 1.96e-18 | 125 | 557 | 74 | 497 | |
| 1.38e-15 | 129 | 557 | 71 | 492 | |
| 8.36e-15 | 129 | 296 | 67 | 228 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.35e-138 | 9 | 571 | 10 | 574 | Crystal structure of VAO-type flavoprotein MtVAO615 at pH 7.5 from Myceliophthora thermophila C1 [Thermothelomyces thermophilus ATCC 42464],6F73_A Crystal structure of VAO-type flavoprotein MtVAO615 at pH 5.0 from Myceliophthora thermophila C1 [Thermothelomyces thermophilus ATCC 42464],6F73_B Crystal structure of VAO-type flavoprotein MtVAO615 at pH 5.0 from Myceliophthora thermophila C1 [Thermothelomyces thermophilus ATCC 42464] |
|
| 2.91e-67 | 29 | 570 | 30 | 597 | Crystal structure of VAO-type flavoprotein MtVAO713 from Myceliophthora thermophila C1 [Thermothelomyces thermophilus ATCC 42464],6F74_B Crystal structure of VAO-type flavoprotein MtVAO713 from Myceliophthora thermophila C1 [Thermothelomyces thermophilus ATCC 42464],6F74_C Crystal structure of VAO-type flavoprotein MtVAO713 from Myceliophthora thermophila C1 [Thermothelomyces thermophilus ATCC 42464],6F74_D Crystal structure of VAO-type flavoprotein MtVAO713 from Myceliophthora thermophila C1 [Thermothelomyces thermophilus ATCC 42464] |
|
| 6.94e-17 | 125 | 547 | 58 | 502 | Crystal structure of tetrahydrocannabinolic acid synthase from Cannabis sativa [Cannabis sativa] |
|
| 7.55e-16 | 129 | 296 | 45 | 206 | Crystal structure of carbohydrate oxidase from Microdochium nivale [Microdochium nivale],3RJA_A Crystal structure of carbohydrate oxidase from Microdochium nivale in complex with substrate analogue [Microdochium nivale] |
|
| 2.76e-15 | 147 | 562 | 78 | 517 | Structure of berberine bridge enzyme in complex with dehydroscoulerine [Eschscholzia californica] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7.13e-183 | 24 | 571 | 22 | 563 | FAD-linked oxidoreductase ZEB1 OS=Gibberella zeae (strain ATCC MYA-4620 / CBS 123657 / FGSC 9075 / NRRL 31084 / PH-1) OX=229533 GN=ZEB1 PE=2 SV=2 |
|
| 4.12e-154 | 26 | 571 | 25 | 567 | FAD-linked oxidoreductase patO OS=Penicillium expansum OX=27334 GN=patO PE=1 SV=1 |
|
| 6.83e-153 | 25 | 570 | 25 | 567 | FAD-linked oxidoreductase patO OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=patO PE=1 SV=1 |
|
| 1.55e-143 | 6 | 570 | 14 | 572 | FAD-linked oxidoreductase sor8 OS=Hypocrea jecorina (strain QM6a) OX=431241 GN=sor8 PE=3 SV=1 |
|
| 2.24e-137 | 9 | 571 | 10 | 574 | VAO-type flavoprotein oxidase VAO615 OS=Myceliophthora thermophila (strain ATCC 42464 / BCRC 31852 / DSM 1799) OX=573729 GN=MYCTH_2305637 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
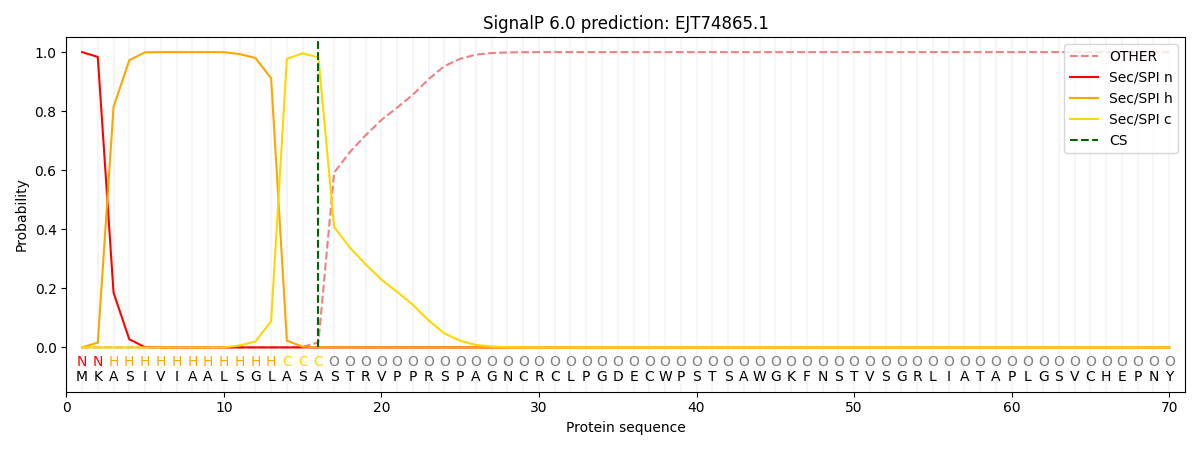
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000325 | 0.999639 | CS pos: 16-17. Pr: 0.9821 |
