You are browsing environment: FUNGIDB
CAZyme Information: EJT72816.1
You are here: Home > Sequence: EJT72816.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Gaeumannomyces tritici | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Magnaporthaceae; Gaeumannomyces; Gaeumannomyces tritici | |||||||||||
| CAZyme ID | EJT72816.1 | |||||||||||
| CAZy Family | GT8 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 227625 | Scw11 | 3.19e-55 | 278 | 551 | 28 | 299 | Exo-beta-1,3-glucanase, GH17 family [Carbohydrate transport and metabolism]. |
| 223021 | PHA03247 | 2.60e-05 | 56 | 301 | 2735 | 2969 | large tegument protein UL36; Provisional |
| 273167 | rad23 | 0.002 | 40 | 108 | 71 | 134 | UV excision repair protein Rad23. All proteins in this family for which functions are known are components of a multiprotein complex used for targeting nucleotide excision repair to specific parts of the genome. In humans, Rad23 complexes with the XPC protein. This family is based on the phylogenomic analysis of JA Eisen (1999, Ph.D. Thesis, Stanford University). [DNA metabolism, DNA replication, recombination, and repair] |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.86e-188 | 1 | 556 | 1 | 539 | |
| 7.73e-154 | 5 | 556 | 4 | 533 | |
| 9.85e-142 | 1 | 556 | 1 | 532 | |
| 9.85e-142 | 1 | 556 | 1 | 532 | |
| 1.99e-131 | 1 | 557 | 1 | 582 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.47e-07 | 327 | 545 | 67 | 278 | Crystal structure of glycoside hydrolase family 17 beta-1,3-glucanosyltransferase from Rhizomucor miehei [Rhizomucor miehei CAU432] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.20e-98 | 1 | 556 | 1 | 564 | Probable beta-glucosidase btgE OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=btgE PE=3 SV=2 |
|
| 4.20e-98 | 1 | 556 | 1 | 564 | Probable beta-glucosidase btgE OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=btgE PE=3 SV=2 |
|
| 4.11e-90 | 123 | 556 | 137 | 551 | Probable beta-glucosidase btgE OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=btgE PE=3 SV=1 |
|
| 1.56e-89 | 1 | 556 | 1 | 564 | Probable beta-glucosidase btgE OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=btgE PE=3 SV=1 |
|
| 2.57e-86 | 1 | 556 | 1 | 601 | Probable beta-glucosidase btgE OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=btgE PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
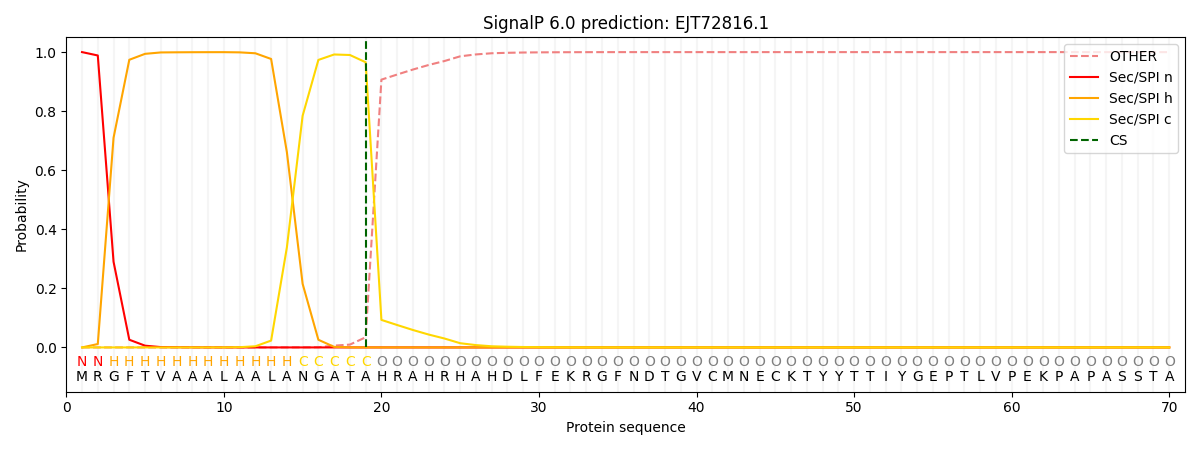
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000718 | 0.999258 | CS pos: 19-20. Pr: 0.9656 |
